Identification of the binding sites of regulatory proteins in bacterial genomes - PubMed (original) (raw)
Identification of the binding sites of regulatory proteins in bacterial genomes
Hao Li et al. Proc Natl Acad Sci U S A. 2002.
Abstract
We present an algorithm that extracts the binding sites (represented by position-specific weight matrices) for many different transcription factors from the regulatory regions of a genome, without the need for delineating groups of coregulated genes. The algorithm uses the fact that many DNA-binding proteins in bacteria bind to a bipartite motif with two short segments more conserved than the intervening region. It identifies all statistically significant patterns of the form W(1)N(x)W(2), where W(1) and W(2) are two short oligonucleotides separated by x arbitrary bases, and groups them into clusters of similar patterns. These clusters are then used to derive quantitative recognition profiles of putative regulatory proteins. For a given cluster, the algorithm finds the matching sequences plus the flanking regions in the genome and performs a multiple sequence alignment to derive position-specific weight matrices. We have analyzed the Escherichia coli genome with this algorithm and found approximately 1,500 significant patterns, which give rise to approximately 160 distinct position-specific weight matrices. A fraction of these matrices match the binding sites of one-third of the approximately 60 characterized transcription factors with high statistical significance. Many of the remaining matrices are likely to describe binding sites and regulons of uncharacterized transcription factors. The significance of these matrices was evaluated by their specificity, the location of the predicted sites, and the biological functions of the corresponding regulons, allowing us to suggest putative regulatory functions. The algorithm is efficient for analyzing newly sequenced bacterial genomes for which little is known about transcriptional regulation.
Figures
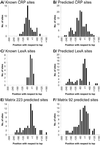
Figure 1
Distribution of the center positions of the predicted binding sites (relative to the transcriptional start point) for four weight matrices. For the two matrices matching CRP and LexA, positional distribution of the known binding sites is also shown.
Similar articles
- Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics.
van Nimwegen E, Zavolan M, Rajewsky N, Siggia ED. van Nimwegen E, et al. Proc Natl Acad Sci U S A. 2002 May 28;99(11):7323-8. doi: 10.1073/pnas.112690399. Proc Natl Acad Sci U S A. 2002. PMID: 12032281 Free PMC article. - Reconstruction of novel transcription factor regulons through inference of their binding sites.
Elmas A, Wang X, Samoilov MS. Elmas A, et al. BMC Bioinformatics. 2015 Sep 21;16:299. doi: 10.1186/s12859-015-0685-y. BMC Bioinformatics. 2015. PMID: 26388177 Free PMC article. - PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny.
Siddharthan R, Siggia ED, van Nimwegen E. Siddharthan R, et al. PLoS Comput Biol. 2005 Dec;1(7):e67. doi: 10.1371/journal.pcbi.0010067. Epub 2005 Dec 9. PLoS Comput Biol. 2005. PMID: 16477324 Free PMC article. - Identifying target sites for cooperatively binding factors.
GuhaThakurta D, Stormo GD. GuhaThakurta D, et al. Bioinformatics. 2001 Jul;17(7):608-21. doi: 10.1093/bioinformatics/17.7.608. Bioinformatics. 2001. PMID: 11448879 - Comparative analysis of regulatory patterns in bacterial genomes.
Gelfand MS, Novichkov PS, Novichkova ES, Mironov AA. Gelfand MS, et al. Brief Bioinform. 2000 Nov;1(4):357-71. doi: 10.1093/bib/1.4.357. Brief Bioinform. 2000. PMID: 11465053 Review.
Cited by
- GeF-seq: A Simple Procedure for Base-Pair Resolution ChIP-seq.
Chumsakul O, Nakamura K, Fukamachi K, Ishikawa S, Oshima T. Chumsakul O, et al. Methods Mol Biol. 2024;2819:39-53. doi: 10.1007/978-1-0716-3930-6_3. Methods Mol Biol. 2024. PMID: 39028501 - Exact p-value calculation for heterotypic clusters of regulatory motifs and its application in computational annotation of cis-regulatory modules.
Boeva V, Clément J, Régnier M, Roytberg MA, Makeev VJ. Boeva V, et al. Algorithms Mol Biol. 2007 Oct 10;2:13. doi: 10.1186/1748-7188-2-13. Algorithms Mol Biol. 2007. PMID: 17927813 Free PMC article. - TRACTOR_DB: a database of regulatory networks in gamma-proteobacterial genomes.
González AD, Espinosa V, Vasconcelos AT, Pérez-Rueda E, Collado-Vides J. González AD, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D98-102. doi: 10.1093/nar/gki054. Nucleic Acids Res. 2005. PMID: 15608293 Free PMC article. - SIGffRid: a tool to search for sigma factor binding sites in bacterial genomes using comparative approach and biologically driven statistics.
Touzain F, Schbath S, Debled-Rennesson I, Aigle B, Kucherov G, Leblond P. Touzain F, et al. BMC Bioinformatics. 2008 Jan 31;9:73. doi: 10.1186/1471-2105-9-73. BMC Bioinformatics. 2008. PMID: 18237374 Free PMC article. - The cis-regulatory map of Shewanella genomes.
Liu J, Xu X, Stormo GD. Liu J, et al. Nucleic Acids Res. 2008 Sep;36(16):5376-90. doi: 10.1093/nar/gkn515. Epub 2008 Aug 13. Nucleic Acids Res. 2008. PMID: 18701645 Free PMC article.
References
- Roth F P, Hughes J D, Estep P W, Church G M. Nat Biotechnol. 1998;16:939–945. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases