Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus - PubMed (original) (raw)
Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus
Sharon J Peacock et al. Infect Immun. 2002 Sep.
Abstract
Most cases of severe Staphylococcus aureus disease cannot be explained by the action of a single virulence determinant, and it is likely that a number of factors act in combination during the infective process. This study examined the relationship between disease in humans and a large number of putative virulence determinants, both individually and in combination. S. aureus isolates (n = 334) from healthy blood donors and from patients with invasive disease were compared for variation in the presence of 33 putative virulence determinants. After adjusting for the effect of clonality, seven determinants (fnbA, cna, sdrE, sej, eta, hlg, and ica) were significantly more common in invasive isolates. All seven factors contributed independently to virulence. No single factor predominated as the major predictor of virulence, their effects appearing to be cumulative. No combinations of the seven genes were either more or less likely to cause disease than others with the same number of virulence-associated genes. There was evidence of considerable horizontal transfer of genes on a background of clonality. Our findings also suggested that allelic variants of a polymorphic locus can make different contributions to the disease process, further study of which is likely to expand our understanding of staphylococcal disease pathogenesis.
Figures
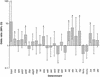
FIG. 1.
Presence or absence of putative virulence determinants. Odds ratios for disease, adjusted for the effects of clonality by Mantel-Haenszel stratification of MLST-defined lineage. Error bars denote 99% confidence intervals (CIs), and an asterisk indicates a significant association with disease (P < 0.01). Genes that were either ubiquitous (such as clfA and clfB) or very rare or absent (such as see and etb) are not shown.
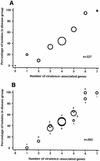
FIG. 2.
(A) Association between the number of virulence-associated determinants and the proportion of isolates from cases of disease (rather than carriage). For example, 20% of isolates with one virulence-associated gene were from cases of disease and 80% were carriage strains. The area of each circle is proportional to the number of isolates with that number of determinants (ranging from zero genes to seven genes, with n = 2, 15, 21, 64, 140, 56, 22, and 7, respectively). (B) Association between the number of virulence-associated determinants in a gene combination and the proportion of isolates with that combination from cases of disease (rather than carriage). Each combination is identified by a letter; for the actual gene complement, refer to Table 4. The 12 most common combinations representing 80% of isolates are shown. The area of each circle is proportional to the number of isolates in that gene combination (n for each gene combination is given in Table 4).
FIG. 3.
Distribution of genes within the seven largest MLST-defined clonal complexes (denoted by numbers along the x axes in the bottom row). The overall height of each bar denotes the total number of isolates in the complex. The height of the shaded area represents the number of isolates positive for the determinant. The population genetic structure of S. aureus can be found on the MLST website (
).
Similar articles
- Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers.
Nashev D, Toshkova K, Salasia SI, Hassan AA, Lämmler C, Zschöck M. Nashev D, et al. FEMS Microbiol Lett. 2004 Apr 1;233(1):45-52. doi: 10.1016/j.femsle.2004.01.032. FEMS Microbiol Lett. 2004. PMID: 15043868 - Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus.
Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Diep BA, et al. J Infect Dis. 2006 Jun 1;193(11):1495-503. doi: 10.1086/503777. Epub 2006 Apr 21. J Infect Dis. 2006. PMID: 16652276 - A link between virulence and ecological abundance in natural populations of Staphylococcus aureus.
Day NP, Moore CE, Enright MC, Berendt AR, Smith JM, Murphy MF, Peacock SJ, Spratt BG, Feil EJ. Day NP, et al. Science. 2001 Apr 6;292(5514):114-6. doi: 10.1126/science.1056495. Science. 2001. PMID: 11292876 Retracted. - Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai.
Song M, Bai Y, Xu J, Carter MQ, Shi C, Shi X. Song M, et al. Int J Food Microbiol. 2015 Feb 16;195:1-8. doi: 10.1016/j.ijfoodmicro.2014.11.020. Epub 2014 Nov 29. Int J Food Microbiol. 2015. PMID: 25485928 - Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus.
Tristan A, Ferry T, Durand G, Dauwalder O, Bes M, Lina G, Vandenesch F, Etienne J. Tristan A, et al. J Hosp Infect. 2007 Jun;65 Suppl 2:105-9. doi: 10.1016/S0195-6701(07)60025-5. J Hosp Infect. 2007. PMID: 17540252 Review.
Cited by
- In silico testing to identify compounds that inhibit ClfA and ClfB binding to the host for the formulation of future drugs against Staphylococcus aureus colonization and infection.
Singh SK, Bhattacharjee M, Unni B, Kashyap RS, Malik A, Akhtar S, Fatima S. Singh SK, et al. Front Cell Infect Microbiol. 2024 Oct 1;14:1422500. doi: 10.3389/fcimb.2024.1422500. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39411322 Free PMC article. - Staphylococcus aureus Co-Infection in COVID-19 Patients: Virulence Genes and Their Influence on Respiratory Epithelial Cells in Light of Risk of Severe Secondary Infection.
Piechowicz L, Kosznik-Kwaśnicka K, Jarzembowski T, Daca A, Necel A, Bonawenturczak A, Werbowy O, Stasiłojć M, Pałubicka A. Piechowicz L, et al. Int J Mol Sci. 2024 Sep 18;25(18):10050. doi: 10.3390/ijms251810050. Int J Mol Sci. 2024. PMID: 39337536 Free PMC article. - Antimicrobial effect of combined preservatives using chestnut inner shell, cinnamon, and ε-poly-lysine against food-poisoning bacteria Staphylococcus aureus.
Lee NK, Park YJ, Kang CE, Kim JH, Shin D, Lee DH, Paik HD. Lee NK, et al. Food Sci Biotechnol. 2024 Jun 25;33(14):3379-3386. doi: 10.1007/s10068-024-01588-y. eCollection 2024 Nov. Food Sci Biotechnol. 2024. PMID: 39328234 - High toxinogenic potential of Staphylococcus aureus from wild ungulates in Brandenburg, Germany with a low level of antibiotic resistance.
Lienen T, Mateus-Vargas RH, Steinhoff-Wagner J, Richter MH, Maurischat S. Lienen T, et al. Front Vet Sci. 2024 Jul 23;11:1445413. doi: 10.3389/fvets.2024.1445413. eCollection 2024. Front Vet Sci. 2024. PMID: 39109350 Free PMC article. - Does Adjunctive Clindamycin Have a Role in Staphylococcus aureus Bacteremia? A Protocol for the Adjunctive Treatment Domain of the Staphylococcus aureus Network Adaptive Platform (SNAP) Randomized Controlled Trial.
Anpalagan K, Dotel R, MacFadden DR, Smith S, Voss L, Petersiel N, Marks M, Marsh J, Mahar RK, McGlothlin A, Lee TC, Goodman A, Morpeth S, Davis JS, Tong SYC, Bowen AC; Adjunctive Clindamycin Domain-Specific Working Group for the Staphylococcus aureus Network Adaptive Platform (SNAP) Trial Group. Anpalagan K, et al. Clin Infect Dis. 2024 Sep 26;79(3):626-634. doi: 10.1093/cid/ciae289. Clin Infect Dis. 2024. PMID: 38801783 Free PMC article. Clinical Trial.
References
- Arbuthnott, J. P. 1982. Bacterial cytolysins (membrane-damaging toxins), p. 107-129. In P. Cohen and S. van Heyningen (ed.), Molecular actions of toxins and viruses. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- Boden, M. K., and J. I. Flock. 1994. Cloning and characterization of a gene for a 19 kDa fibrinogen-binding protein from Staphylococcus aureus. Mol. Microbiol. 12:599-606. - PubMed
- Boden, M. K., and J. I. Flock. 1992. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb. Pathog. 12:289-298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous