Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator - PubMed (original) (raw)
Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator
Sailesh Surapureddi et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Peroxisome proliferator-activated receptor alpha (PPAR alpha) plays a central role in the cell-specific pleiotropic responses induced by structurally diverse synthetic chemicals designated as peroxisome proliferators. Transcriptional regulation by liganded nuclear receptors involves the participation of cofactors that form multiprotein complexes to achieve cell- and gene-specific transcription. Here we report the identification of such a transcriptionally active PPAR alpha-interacting cofactor (PRIC) complex from rat liver nuclear extracts that interacts with full-length PPAR alpha in the presence of ciprofibrate, a synthetic ligand, and leukotriene B(4), a natural ligand. The liganded PPAR alpha-PRIC complex enhanced transcription from a peroxisomal enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme gene promoter template that contains peroxisome proliferator response elements. Rat liver PRIC complex comprises some 25 polypeptides, and their identities were established by mass spectrometry and limited sequence analysis. Eighteen of these peptides contain one or more LXXLL motifs necessary for interacting with nuclear receptors. PRIC complex includes known coactivators or coactivator-binding proteins (CBP, SRC-1, PBP, PRIP, PIMT, TRAP100, SUR-2, and PGC-1), other proteins that have not previously been described in association with transcription complexes (CHD5, TOG, and MORF), and a few novel polypeptides designated PRIC300, -285, -215, -177, and -145. We describe the cDNA for PRIC285, which contains five LXXLL motifs. It interacts with PPAR alpha and acts as a coactivator by moderately stimulating PPAR alpha-mediated transcription in transfected cells. We conclude that liganded PPAR alpha recruits a distinctive multiprotein complex from rat liver nuclear extracts. The composition of this complex may provide insight into the basis of tissue and species sensitivity to peroxisome proliferators.
Figures
Figure 1
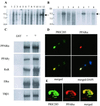
Purification and identification of proteins interacting with PPARα. (A) Identification of ligand-dependent PPARα-interacting complex. SDS/PAGE analysis and silver staining of proteins bound to GST alone (lane 1), GST-PPARα in the absence of ligand (lane 2), and GST-PPARα in the presence of natural ligand LTB4 (lane 3) or synthetic ligand ciprofibrate (Cip, lane 4). (B) Mass spectrometric and limited sequence analysis data. PRIC subunit number, peptide sequence(s), experimental monoisotopic mass (m/z), SwissProt database accession number(s), and identity are included. Of the 25 PRIC peptides, 18 (bold-faced) have one or more LXXLL motifs.
Figure 2
Purified rat nuclear PRIC potentiates PPARα transcription in a cell-free system. In vitro transcription reactions were set up as described in Materials and Methods. A 3.6-kb promoter of the rat L-PBE gene served as template for the transcription assay, with minimal nuclear extract as the source for basal transcription factors and DNA-dependent RNA polymerase II. Purified PPARα or RXR protein was added to the template DNA before the addition of nuclear extract and other components. The reactions were allowed to proceed for 60 min at 30°C, and labeled RNA transcript was extracted and separated on a urea/TBE gel and autoradiographed. GST eluate and control eluate represent, respectively, fraction bound to GST alone and to GST-PPARα in the absence of added ligand. GSH and Sarkosyl eluates represent proteins bound to GST-PPARα in the presence of ligand.
Figure 3
Deduced human PRIC285 amino acid sequence with five LXXLL motifs (boldfaced). Two peptide sequences found by mass spectrometry are underlined. The putative UvrD/helicase domain is between amino acids 1584 and 1991.
Figure 4
Northern blotting, binding of PRIC285 to PPARα, and colocalization. (A) Northern analysis of human multiple tissue blot (CLONTECH). Lanes 1–12 represent, respectively, brain, heart, skeletal muscle, colon, thymus, spleen, kidney, liver, small intestine, placenta, lung, and leukocytes. Arrow indicates PRIC285 mRNA. (B) Northern analysis of human cancer cell line blot (CLONTECH) probed with a partial PRIC285 cDNA probe. Lanes 1–8 represent, respectively, HL-60 cells line, HeLa cell S3, chronic myelogenous leukemia (K-562), lymphoblastic leukemia MOLT-4, Burkitt's lymphoma Raji, colorectal adenocarcinoma SW480, lung carcinoma A-549, and melanoma G-361. Arrow indicates PRIC285 mRNA. (C) [35S]Methionine-labeled full-length PRIC285 generated by in vitro translation was incubated with GSH-Sepharose beads bound with purified _E. coli_-expressed GST-PPARα or GST. PRIC285 bound to PPARα in the absence of ligand, and the addition of ligand increased the binding ≈2-fold. PRIC285 also interacts with PPARγ, RXRα, ERα (estrogen receptor α), and TRβ1 (thyroid hormone receptor β1). (D and E) Colocalization of PRIC285 and PPARα in the nucleus as visualized by conventional fluorescence microscopy (D) and by DELTAVISION deconvolution microscopy (E). PRIC285 was expressed transiently in HEK293 cells by using three FLAG epitopes linked to the C terminus, and it was visualized by using anti-FLAG antibodies (green). PPARα was also transiently expressed in these cells and localized by using anti-PPARα antibodies (red). Merging of the PRIC285 and PPARα localization images of the same cell reveals overlapping localization (yellow) of these two proteins. 4×,6-Diamidino-2-phenylindole (DAPI) staining shows nuclei in D (Lower Right).
Figure 5
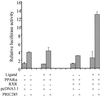
PRIC285 functions as a coactivator for PPARα. HEK293 cells were cotransfected with 1.5 μg of reporter construct PPRE-TK-LUC, 0.25 μg of PCMV-PPARα, 0.25 μg of PCMV-RXR, 0.5 μg of PCMV-PRIC285, and 0.5 μg of pcDNA3.1 as indicated. Transfections without the indicated plasmid were compensated by adding the same amount of pcDNA3.1. The activity obtained on transfection of PPRE-TK-LUC without exogenous PRIC285 was taken as 1. Results are the mean of three independent transfections.
Similar articles
- PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex.
Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. Surapureddi S, et al. Biochem Biophys Res Commun. 2006 May 5;343(2):535-43. doi: 10.1016/j.bbrc.2006.02.160. Epub 2006 Mar 9. Biochem Biophys Res Commun. 2006. PMID: 16554032 - Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver.
Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, Le Roith D, Chambon P, Gonzalez FJ, Reddy JK. Jia Y, et al. J Biol Chem. 2004 Jun 4;279(23):24427-34. doi: 10.1074/jbc.M402391200. Epub 2004 Mar 29. J Biol Chem. 2004. PMID: 15150259 - Peroxisome proliferator-activated receptors, coactivators, and downstream targets.
Qi C, Zhu Y, Reddy JK. Qi C, et al. Cell Biochem Biophys. 2000;32 Spring:187-204. doi: 10.1385/cbb:32:1-3:187. Cell Biochem Biophys. 2000. PMID: 11330046 Review. - Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression.
Marcus SL, Miyata KS, Zhang B, Subramani S, Rachubinski RA, Capone JP. Marcus SL, et al. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5723-7. doi: 10.1073/pnas.90.12.5723. Proc Natl Acad Sci U S A. 1993. PMID: 8390676 Free PMC article. - Regulation of the peroxisomal beta-oxidation-dependent pathway by peroxisome proliferator-activated receptor alpha and kinases.
Latruffe N, Cherkaoui Malki M, Nicolas-Frances V, Clemencet MC, Jannin B, Berlot JP. Latruffe N, et al. Biochem Pharmacol. 2000 Oct 15;60(8):1027-32. doi: 10.1016/s0006-2952(00)00416-0. Biochem Pharmacol. 2000. PMID: 11007938 Review.
Cited by
- A rare nonsynonymous variant in the lipid metabolic gene HELZ2 related to primary biliary cirrhosis in Chinese Han.
Li P, Lu G, Wang L, Cui Y, Wu Z, Chen S, Li J, Wen X, Zhang H, Mu S, Zhang F, Li Y. Li P, et al. Allergy Asthma Clin Immunol. 2016 Apr 4;12:14. doi: 10.1186/s13223-016-0120-6. eCollection 2016. Allergy Asthma Clin Immunol. 2016. PMID: 27047549 Free PMC article. - HELZ2 Is an IFN Effector Mediating Suppression of Dengue Virus.
Fusco DN, Pratt H, Kandilas S, Cheon SS, Lin W, Cronkite DA, Basavappa M, Jeffrey KL, Anselmo A, Sadreyev R, Yapp C, Shi X, O'Sullivan JF, Gerszten RE, Tomaru T, Yoshino S, Satoh T, Chung RT. Fusco DN, et al. Front Microbiol. 2017 Feb 20;8:240. doi: 10.3389/fmicb.2017.00240. eCollection 2017. Front Microbiol. 2017. PMID: 28265266 Free PMC article. - Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases.
Olivares AM, Moreno-Ramos OA, Haider NB. Olivares AM, et al. J Exp Neurosci. 2016 May 5;9(Suppl 2):93-121. doi: 10.4137/JEN.S25480. eCollection 2015. J Exp Neurosci. 2016. PMID: 27168725 Free PMC article. Review. - Non-DNA binding, dominant-negative, human PPARgamma mutations cause lipodystrophic insulin resistance.
Agostini M, Schoenmakers E, Mitchell C, Szatmari I, Savage D, Smith A, Rajanayagam O, Semple R, Luan J, Bath L, Zalin A, Labib M, Kumar S, Simpson H, Blom D, Marais D, Schwabe J, Barroso I, Trembath R, Wareham N, Nagy L, Gurnell M, O'Rahilly S, Chatterjee K. Agostini M, et al. Cell Metab. 2006 Oct;4(4):303-11. doi: 10.1016/j.cmet.2006.09.003. Cell Metab. 2006. PMID: 17011503 Free PMC article. - Influence of Varying Dietary ω6 to ω3 Fatty Acid Ratios on the Hepatic Transcriptome, and Association with Phenotypic Traits (Growth, Somatic Indices, and Tissue Lipid Composition), in Atlantic Salmon (Salmo salar).
Katan T, Xue X, Caballero-Solares A, Taylor RG, Parrish CC, Rise ML. Katan T, et al. Biology (Basel). 2021 Jun 24;10(7):578. doi: 10.3390/biology10070578. Biology (Basel). 2021. PMID: 34202562 Free PMC article.
References
- Reddy J K, Krishnakantha T P. Science. 1975;190:787–789. - PubMed
- Reddy J K, Chu R. Ann NY Acad Sci. 1996;804:176–201. - PubMed
- Issemann J, Green S. Nature (London) 1990;347:645–650. - PubMed
- Gonzalez F J, Peters J M, Cattley R C. J Natl Cancer Inst. 1998;90:1702–1709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM23570/GM/NIGMS NIH HHS/United States
- CA84472/CA/NCI NIH HHS/United States
- R37 GM023750/GM/NIGMS NIH HHS/United States
- R01 CA084472/CA/NCI NIH HHS/United States
- R01 GM023750/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous