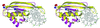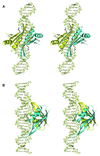Structure of Alba: an archaeal chromatin protein modulated by acetylation - PubMed (original) (raw)
Comparative Study
Structure of Alba: an archaeal chromatin protein modulated by acetylation
B N Wardleworth et al. EMBO J. 2002.
Abstract
Eukaryotic DNA is packaged into nucleosomes that regulate the accessibility of the genome to replication, transcription and repair factors. Chromatin accessibility is controlled by histone modifications including acetylation and methylation. Archaea possess eukary otic-like machineries for DNA replication, transcription and information processing. The conserved archaeal DNA binding protein Alba (formerly Sso10b) interacts with the silencing protein Sir2, which regulates Alba's DNA binding affinity by deacetylation of a lysine residue. We present the crystal structure of Alba from Sulfolobus solfataricus at 2.6 A resolution (PDB code 1h0x). The fold is reminiscent of the N-terminal DNA binding domain of DNase I and the C-terminal domain of initiation factor IF3. The Alba dimer has two extended beta-hairpins flanking a central body containing the acetylated lysine, Lys16, suggesting three main points of contact with the DNA. Fluorescence, calorimetry and electrophoresis data suggest a final binding stoichiometry of approximately 5 bp DNA per Alba dimer. We present a model for the Alba-DNA interaction consistent with the available structural, biophysical and electron microscopy data.
Figures
Fig. 1. Sequence alignment of archaeal Alba homologues. More divergent homologues of Alba exist in trypanosomes, higher plants and vertebrates (Bell et al., 2002). Sso (S.solfataricus P74761), Pae (Pyrobaculum aerophilum Q8ZVL3), Mja (Methanococcus jannaschii Q57665), Pab (Pyrococcus abyssi Q9VIN3), Afu (Archaeoglobus fulgidus O28323), Mth (Methanothermobacter thermautotrophicus O27527), Tac (Thermoplasma acidophilum Q9HJQ5) are aligned. The secondary structure of Sulfolobus Alba derived from the crystal structure is shown above the alignment. Residues involved in the main dimer interface are marked with a ‘+’.
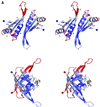
Fig. 2. The Alba dimer. (A) Stereo views of the dimer coloured by _B_-factor (dark blue to deep red representing a span of _B_-factor from 30 to 100 Å2). The lower view is related to the upper view by a 90° rotation around a vertical axis. The strands and helices of one monomer are labelled as in Figure 1. The N- and C-termini are highlighted by blue and red spheres, respectively. Lysines 16 and 17 are also shown. (B) Orthogonal views of the dimer showing the location of exposed residues conserved across the Archaea: A (Gly15, Lys17, Pro18, Asn21, Tyr22), B (Lys40, Arg42, Glu91) and C (Phe60). (C) Stereo view of the Alba dimer coloured by electrostatic potential. The groove formed between the two loops containing Lys16 and Lys17 is apparent. Figures 2, 3 and 5 were drawn with BOBSCRIPT (Esnouf, 1997) and GL_RENDER (L.Esser and J.Deisenhofer, unpublished).
Fig. 2. The Alba dimer. (A) Stereo views of the dimer coloured by _B_-factor (dark blue to deep red representing a span of _B_-factor from 30 to 100 Å2). The lower view is related to the upper view by a 90° rotation around a vertical axis. The strands and helices of one monomer are labelled as in Figure 1. The N- and C-termini are highlighted by blue and red spheres, respectively. Lysines 16 and 17 are also shown. (B) Orthogonal views of the dimer showing the location of exposed residues conserved across the Archaea: A (Gly15, Lys17, Pro18, Asn21, Tyr22), B (Lys40, Arg42, Glu91) and C (Phe60). (C) Stereo view of the Alba dimer coloured by electrostatic potential. The groove formed between the two loops containing Lys16 and Lys17 is apparent. Figures 2, 3 and 5 were drawn with BOBSCRIPT (Esnouf, 1997) and GL_RENDER (L.Esser and J.Deisenhofer, unpublished).
Fig. 3. Comparison of DNase I with Alba. The N-terminal domain, residues 1–86, of DNase I is coloured magenta, in complex with a nicked DNA octamer (PDB code 2DNJ), and showing the β-hairpin that interacts with the DNA minor groove. An Alba monomer is superimposed in yellow, revealing the more extensive β-hairpin of Alba, and suggesting that the orientation of the DNA will be different in the Alba–DNA complex. The phosphorus atoms of the DNA are coloured green, and the side-chains of lysines 16 and 17 of Alba are shown.
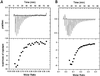
Fig. 4. Interaction of Alba with DNA. (A and B) ITC. Titration of recombinant Alba into supercoiled pUC18 plasmid DNA at 50°C (A) and 25°C (B). Upper panels: raw data for sequential 10 µl injections of 300 µM recombinant Alba into a solution containing 0.05 µM pUC18 plasmid. Lower panel: integrated heat data. Exothermic binding was observed at both temperatures, with a complex binding curve that could not be fitted using standard models. The _x_-axis denotes the stoichiometry of Alba dimers per base pair of duplex DNA. Saturation of the DNA binding sites was evident at between 10 and 5 bp dsDNA per dimer of Alba. (C) DAPI dye displacement assay. Supercoiled phiX174 plasmid (1 nM), saturated with the fluorescent dye DAPI in binding buffer was incubated with increasing concentrations of recombinant Alba protein, and the decrease in fluorescence intensity due to displacement of the DAPI was monitored as described in Materials and methods. Completion of the dye displacement was evident at a ratio of between 10 and 5 bp dsDNA per dimer of Alba. (D) Agarose gel electrophoresis. Two hundred and fifty nanograms of supercoiled phiX174 dsDNA (sc) were incubated with increasing concentrations of native (acetylated) or recombinant (unacetylated) Alba prior to electrophoresis in a 0.7% agarose/1× TBE gel. The molar ratio of (base pairs dsDNA to Alba dimers) is indicated. Decreases in the DNA mobility are evident at 20 bp per dimer for both forms of the protein, with the most significant shift occurring between 10 and 7.5 bp per dimer. There is little obvious difference between the stoichiometry of DNA binding of the native and recombinant protein, despite the 30-fold lower binding affinity of the former. The small amount of nicked, open circle (oc) DNA present in the sample is retarded similarly to the supercoiled form (sc).
Fig. 4. Interaction of Alba with DNA. (A and B) ITC. Titration of recombinant Alba into supercoiled pUC18 plasmid DNA at 50°C (A) and 25°C (B). Upper panels: raw data for sequential 10 µl injections of 300 µM recombinant Alba into a solution containing 0.05 µM pUC18 plasmid. Lower panel: integrated heat data. Exothermic binding was observed at both temperatures, with a complex binding curve that could not be fitted using standard models. The _x_-axis denotes the stoichiometry of Alba dimers per base pair of duplex DNA. Saturation of the DNA binding sites was evident at between 10 and 5 bp dsDNA per dimer of Alba. (C) DAPI dye displacement assay. Supercoiled phiX174 plasmid (1 nM), saturated with the fluorescent dye DAPI in binding buffer was incubated with increasing concentrations of recombinant Alba protein, and the decrease in fluorescence intensity due to displacement of the DAPI was monitored as described in Materials and methods. Completion of the dye displacement was evident at a ratio of between 10 and 5 bp dsDNA per dimer of Alba. (D) Agarose gel electrophoresis. Two hundred and fifty nanograms of supercoiled phiX174 dsDNA (sc) were incubated with increasing concentrations of native (acetylated) or recombinant (unacetylated) Alba prior to electrophoresis in a 0.7% agarose/1× TBE gel. The molar ratio of (base pairs dsDNA to Alba dimers) is indicated. Decreases in the DNA mobility are evident at 20 bp per dimer for both forms of the protein, with the most significant shift occurring between 10 and 7.5 bp per dimer. There is little obvious difference between the stoichiometry of DNA binding of the native and recombinant protein, despite the 30-fold lower binding affinity of the former. The small amount of nicked, open circle (oc) DNA present in the sample is retarded similarly to the supercoiled form (sc).
Fig. 4. Interaction of Alba with DNA. (A and B) ITC. Titration of recombinant Alba into supercoiled pUC18 plasmid DNA at 50°C (A) and 25°C (B). Upper panels: raw data for sequential 10 µl injections of 300 µM recombinant Alba into a solution containing 0.05 µM pUC18 plasmid. Lower panel: integrated heat data. Exothermic binding was observed at both temperatures, with a complex binding curve that could not be fitted using standard models. The _x_-axis denotes the stoichiometry of Alba dimers per base pair of duplex DNA. Saturation of the DNA binding sites was evident at between 10 and 5 bp dsDNA per dimer of Alba. (C) DAPI dye displacement assay. Supercoiled phiX174 plasmid (1 nM), saturated with the fluorescent dye DAPI in binding buffer was incubated with increasing concentrations of recombinant Alba protein, and the decrease in fluorescence intensity due to displacement of the DAPI was monitored as described in Materials and methods. Completion of the dye displacement was evident at a ratio of between 10 and 5 bp dsDNA per dimer of Alba. (D) Agarose gel electrophoresis. Two hundred and fifty nanograms of supercoiled phiX174 dsDNA (sc) were incubated with increasing concentrations of native (acetylated) or recombinant (unacetylated) Alba prior to electrophoresis in a 0.7% agarose/1× TBE gel. The molar ratio of (base pairs dsDNA to Alba dimers) is indicated. Decreases in the DNA mobility are evident at 20 bp per dimer for both forms of the protein, with the most significant shift occurring between 10 and 7.5 bp per dimer. There is little obvious difference between the stoichiometry of DNA binding of the native and recombinant protein, despite the 30-fold lower binding affinity of the former. The small amount of nicked, open circle (oc) DNA present in the sample is retarded similarly to the supercoiled form (sc).
Fig. 5. Models of Alba binding to DNA. (A and B) Orthogonal stereo views of how one Alba dimer might bind to DNA, with the β-hairpins interacting with the minor grooves and the critical Lys16 sitting either side of the duplex. Residues Lys16, Lys17 and Arg42 are highlighted. (C and D) Orthogonal views of a model for the binding of multiple Alba dimers along the DNA duplex. Consecutive dimers are rotated by 120° with respect to one another, allowing close packing of Alba molecules along the DNA duplex with minimal steric clash.
Fig. 5. Models of Alba binding to DNA. (A and B) Orthogonal stereo views of how one Alba dimer might bind to DNA, with the β-hairpins interacting with the minor grooves and the critical Lys16 sitting either side of the duplex. Residues Lys16, Lys17 and Arg42 are highlighted. (C and D) Orthogonal views of a model for the binding of multiple Alba dimers along the DNA duplex. Consecutive dimers are rotated by 120° with respect to one another, allowing close packing of Alba molecules along the DNA duplex with minimal steric clash.
Similar articles
- Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms.
Chou CC, Lin TW, Chen CY, Wang AH. Chou CC, et al. J Bacteriol. 2003 Jul;185(14):4066-73. doi: 10.1128/JB.185.14.4066-4073.2003. J Bacteriol. 2003. PMID: 12837780 Free PMC article. - The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation.
Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. Bell SD, et al. Science. 2002 Apr 5;296(5565):148-51. doi: 10.1126/science.1070506. Science. 2002. PMID: 11935028 - Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding.
Zhao K, Chai X, Marmorstein R. Zhao K, et al. J Biol Chem. 2003 Jul 11;278(28):26071-7. doi: 10.1074/jbc.M303666200. Epub 2003 May 1. J Biol Chem. 2003. PMID: 12730210 - The Alba protein family: Structure and function.
Goyal M, Banerjee C, Nag S, Bandyopadhyay U. Goyal M, et al. Biochim Biophys Acta. 2016 May;1864(5):570-83. doi: 10.1016/j.bbapap.2016.02.015. Epub 2016 Feb 17. Biochim Biophys Acta. 2016. PMID: 26900088 Review. - Archaeal chromatin and transcription.
Reeve JN. Reeve JN. Mol Microbiol. 2003 May;48(3):587-98. doi: 10.1046/j.1365-2958.2003.03439.x. Mol Microbiol. 2003. PMID: 12694606 Review.
Cited by
- Thermal and conformational stability of Ssh10b protein from archaeon Sulfolobus shibattae.
Xu S, Qin S, Pan XM. Xu S, et al. Biochem J. 2004 Sep 1;382(Pt 2):433-40. doi: 10.1042/BJ20040191. Biochem J. 2004. PMID: 15107015 Free PMC article. - Crystal structure of the hyperthermophilic archaeal DNA-binding protein Sso10b2 at a resolution of 1.85 Angstroms.
Chou CC, Lin TW, Chen CY, Wang AH. Chou CC, et al. J Bacteriol. 2003 Jul;185(14):4066-73. doi: 10.1128/JB.185.14.4066-4073.2003. J Bacteriol. 2003. PMID: 12837780 Free PMC article. - ALBA protein complex reads genic R-loops to maintain genome stability in Arabidopsis.
Yuan W, Zhou J, Tong J, Zhuo W, Wang L, Li Y, Sun Q, Qian W. Yuan W, et al. Sci Adv. 2019 May 15;5(5):eaav9040. doi: 10.1126/sciadv.aav9040. eCollection 2019 May. Sci Adv. 2019. PMID: 31106272 Free PMC article. - Mth10b, a unique member of the Sac10b family, does not bind nucleic acid.
Liu YF, Zhang N, Yao HW, Pan XM, Ge M. Liu YF, et al. PLoS One. 2011;6(5):e19977. doi: 10.1371/journal.pone.0019977. Epub 2011 May 18. PLoS One. 2011. PMID: 21625642 Free PMC article. - Identification and molecular characterization of an Alba-family protein from human malaria parasite Plasmodium falciparum.
Goyal M, Alam A, Iqbal MS, Dey S, Bindu S, Pal C, Banerjee A, Chakrabarti S, Bandyopadhyay U. Goyal M, et al. Nucleic Acids Res. 2012 Feb;40(3):1174-90. doi: 10.1093/nar/gkr821. Epub 2011 Oct 17. Nucleic Acids Res. 2012. PMID: 22006844 Free PMC article.
References
- Agback P., Baumann,H., Knapp,S., Ladenstein,R. and Hard,T. (1998) Architecture of nonspecific protein–DNA interactions in the Sso7d–DNA complex. Nat. Struct. Biol., 5, 579–584. - PubMed
- Bell S.D. and Jackson,S.P. (1998) Transcription in Archaea. Cold Spring Harb. Symp. Quant. Biol., 63, 41–51. - PubMed
- Bell S.D., Botting,C.H., Wardleworth,B.N., Jackson,S.P. and White,M.F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science, 296, 148–151. - PubMed
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous