Identification of benzothiazoles as potential polyglutamine aggregation inhibitors of Huntington's disease by using an automated filter retardation assay - PubMed (original) (raw)
. 2002 Dec 10;99 Suppl 4(Suppl 4):16400-6.
doi: 10.1073/pnas.182426599. Epub 2002 Aug 28.
Sabine Engemann, Wolfgang Bröcker, Ilona Dunkel, Annett Boeddrich, Stephanie Waelter, Eddi Nordhoff, Rudi Lurz, Nancy Schugardt, Susanne Rautenberg, Christian Herhaus, Gerhard Barnickel, Henning Böttcher, Hans Lehrach, Erich E Wanker
Affiliations
- PMID: 12200548
- PMCID: PMC139900
- DOI: 10.1073/pnas.182426599
Identification of benzothiazoles as potential polyglutamine aggregation inhibitors of Huntington's disease by using an automated filter retardation assay
Volker Heiser et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Preventing the formation of insoluble polyglutamine containing protein aggregates in neurons may represent an attractive therapeutic strategy to ameliorate Huntington's disease (HD). Therefore, the ability to screen for small molecules that suppress the self-assembly of huntingtin would have potential clinical and significant research applications. We have developed an automated filter retardation assay for the rapid identification of chemical compounds that prevent HD exon 1 protein aggregation in vitro. Using this method, a total of 25 benzothiazole derivatives that inhibit huntingtin fibrillogenesis in a dose-dependent manner were discovered from a library of approximately 184,000 small molecules. The results obtained by the filter assay were confirmed by immunoblotting, electron microscopy, and mass spectrometry. Furthermore, cell culture studies revealed that 2-amino-4,7-dimethyl-benzothiazol-6-ol, a chemical compound similar to riluzole, significantly inhibits HD exon 1 aggregation in vivo. These findings may provide the basis for a new therapeutic approach to prevent the accumulation of insoluble protein aggregates in Huntington's disease and related glutamine repeat disorders.
Figures
Figure 1
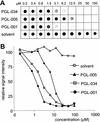
Development of an automated filter retardation assay for high-throughput drug screening. (A) Flow chart of the automated filter retardation assay. (B) Effect of chemical compounds on HD exon 1 aggregation as monitored by the filter assay. GST-HD51 fusion protein at a conc. of 1.25 μM was predigested for 3 min at 37°C with elastase to remove totally the GST tag from the fusion protein and to initiate aggregation of the HDQ51 protein. Immediately after this step, various chemical compounds were added (final conc. 10 μM) and samples were incubated for an additional 16 h at 37°C. Then, aggregation reactions were filtered through a cellulose acetate membrane by using a 384-well dot blot apparatus. Captured aggregates were detected by immunoblotting using the HD1 antibody. On each filter membrane, 320 different chemical compounds were tested (squares A2–H11). Squares A1–H1 and A12–H12 were used for control samples. In square F8, an inhibitory compound was identified. ThioS, thioflavine S.
Figure 2
In vitro inhibition of HD exon 1 aggregation by benzothiazoles. (A) Effect of the indicated chemical compounds on HD exon 1 aggregation as monitored by the filter retardation assay. (B) Quantification of the dot blot results shown in A. The signal intensity from the sample without added chemical compound (solvent) was arbitrarily set as 100. For structure of the chemical compounds, see Table 1.
Figure 3
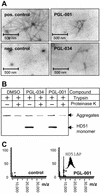
Analysis of HD exon 1 aggregation in vitro by using secondary assays. (A) Electron micrographs of HD51 fibrils formed in the presence or absence of the indicated benzothiazoles. Trypsin-digested GST-HD51 protein at 1.5 μM was incubated for 16 h at 37°C either without chemical compound (pos. control) or with PGL-001 and PGL-034 (final conc. 20 μM). Representative examples of fibrillar structures are shown. (B) Western blot analysis of trypsin-digested GST-HD51 fusion protein. Aggregation reactions were performed for 16 h in the presence or absence of the indicated chemical compounds. Where indicated (+), aliquots were taken and incubated for additional 30 min with 10 ng/μl proteinase K. Samples corresponding to 200 ng of fusion protein were analyzed by SDS/PAGE and immunoblotting using the HD1 antibody. (C) Effect of the compound PGL-001 on HD exon 1 aggregation as monitored by MALDI-TOF MS. GST-HD51ΔP at 2.5 μM was incubated for 18 h at 37°C with trypsin in the absence (control) or presence of PGL-001 (final conc. 20 μM). Analysis by MALDI-TOF MS revealed that, only in the presence of PGL-001, monomeric HD51ΔP peptide (monoisotopic mass of 8,074.74 Da) is detectable; without added compound, the peptide has assembled into insoluble high-molecular-weight protein aggregates that cannot be detected by MALDI-TOF MS.
Figure 4
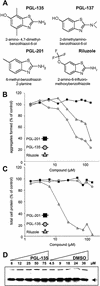
Inhibition of HD exon 1 aggregation in 293 Tet-Off cells. (A) Structure of chemical compounds counteracting HDQ51 aggregation in vivo. (B) Quantification of filter retardation assay results. Cells were incubated for 72 h in the presence of various concentrations of the indicated chemical compounds. Protein extracts were prepared and filtered through a cellulose acetate membrane; captured SDS-insoluble protein aggregates were detected with the HD1 antibody. The dots corresponding to the control reactions without added chemical compound (not shown) were arbitrarily set as 100. (C) Relative protein concentrations of the cell extracts analyzed in B. (D) Western blot of cell extracts prepared from 293 Tet-Off cells after treatment with increasing concentrations of the chemical compound PGL-135 and the solvent DMSO. The arrow indicates the HDQ51 monomer.
Figure 5
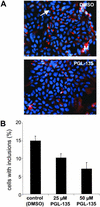
Immunofluorescence microscopy analysis. (A) 293 Tet-Off cells expressing Flag-tagged HD exon 1 protein with 51 glutamines (HDQ51) were cultivated for 72 h in the presence or absence of PGL-135. Formation of inclusion bodies with aggregated HDQ51 protein was followed by indirect immunofluorescence microscopy using the anti-Flag antibody. Inclusion bodies are indicated as red dots by the arrow. Nuclei were counterstained with Hoechst. A total of about 5,000 PGL-135-treated and untreated cells were examined. (B) Quantification of cells with inclusion bodies.
Similar articles
- Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington's disease therapy.
Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker EE. Heiser V, et al. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6739-44. doi: 10.1073/pnas.110138997. Proc Natl Acad Sci U S A. 2000. PMID: 10829068 Free PMC article. - Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models.
Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, Legleiter J, Marsh JL, Thompson LM, Lindquist S, Muchowski PJ, Wanker EE. Ehrnhoefer DE, et al. Hum Mol Genet. 2006 Sep 15;15(18):2743-51. doi: 10.1093/hmg/ddl210. Epub 2006 Aug 7. Hum Mol Genet. 2006. PMID: 16893904 - Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease.
Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, Hayer-Hartl MK, Hartl FU, Wanker EE. Sittler A, et al. Hum Mol Genet. 2001 Jun 1;10(12):1307-15. doi: 10.1093/hmg/10.12.1307. Hum Mol Genet. 2001. PMID: 11406612 - Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease.
Truant R, Atwal RS, Burtnik A. Truant R, et al. Prog Neurobiol. 2007 Nov;83(4):211-27. doi: 10.1016/j.pneurobio.2006.11.004. Epub 2007 Jan 22. Prog Neurobiol. 2007. PMID: 17240517 Review. - Proteomic Analysis of Huntington's Disease.
Kumar S, Singh P, Sharma S, Ali J, Baboota S, Pottoo FH. Kumar S, et al. Curr Protein Pept Sci. 2020;21(12):1218-1222. doi: 10.2174/1389203721666201006160327. Curr Protein Pept Sci. 2020. PMID: 33023443 Review.
Cited by
- Target- and mechanism-based therapeutics for neurodegenerative diseases: strength in numbers.
Trippier PC, Jansen Labby K, Hawker DD, Mataka JJ, Silverman RB. Trippier PC, et al. J Med Chem. 2013 Apr 25;56(8):3121-47. doi: 10.1021/jm3015926. Epub 2013 Mar 27. J Med Chem. 2013. PMID: 23458846 Free PMC article. Review. - Neuroprotective effects of the antioxidant action of 2-cyclopropylimino-3-methyl-1,3-thiazoline hydrochloride against ischemic neuronal damage in the brain.
Ha SC, Han AR, Kim DW, Kim EA, Kim DS, Choi SY, Cho SW. Ha SC, et al. BMB Rep. 2013 Jul;46(7):370-5. doi: 10.5483/bmbrep.2013.46.7.018. BMB Rep. 2013. PMID: 23884104 Free PMC article. - Modified single-stranded oligonucleotides inhibit aggregate formation and toxicity induced by expanded polyglutamine.
Parekh-Olmedo H, Wang J, Gusella JF, Kmiec EB. Parekh-Olmedo H, et al. J Mol Neurosci. 2004;24(2):257-67. doi: 10.1385/JMN:24:2:257. J Mol Neurosci. 2004. PMID: 15456939 - The Aggregation of Huntingtin and α-Synuclein.
Chánez-Cárdenas ME, Vázquez-Contreras E. Chánez-Cárdenas ME, et al. J Biophys. 2012;2012:606172. doi: 10.1155/2012/606172. Epub 2012 Jul 26. J Biophys. 2012. PMID: 22899913 Free PMC article. - NO synthase 2 (NOS2) deletion promotes multiple pathologies in a mouse model of Alzheimer's disease.
Colton CA, Vitek MP, Wink DA, Xu Q, Cantillana V, Previti ML, Van Nostrand WE, Weinberg JB, Dawson H. Colton CA, et al. Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12867-72. doi: 10.1073/pnas.0601075103. Epub 2006 Aug 14. Proc Natl Acad Sci U S A. 2006. PMID: 16908860 Free PMC article.
References
- Harper P S. Huntington's Disease. London: Saunders; 1991.
- Sathasivam K, Amaechi I, Mangiarini L, Bates G. Hum Genet. 1997;99:692–695. - PubMed
- Wanker E E. Biol Chem. 2000;381:937–942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical