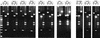Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates - PubMed (original) (raw)
Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates
Patrizia Spigaglia et al. J Clin Microbiol. 2002 Sep.
Abstract
The pathogenicity locus (PaLoc) of Clostridium difficile contains toxin A and B genes and three accessory genes, including tcdD and tcdC, which are supposed to code for the positive and negative regulators of toxin expression, respectively. Different studies have described variations in C. difficile toxin A and B genes, but little is known about C. difficile variants for the accessory genes. The PaLoc of several C. difficile clinical isolates was investigated by three different PCR methods with the aim to identify variant strains. Of the toxinogenic C. difficile strains examined, 25% showed variations. No correlation between C. difficile variant strains and key patient groups was found. Interestingly, all of these strains showed a variant tcdC gene. Three different tcdC alleles were identified, and one of these had a nonsense mutation which reduced the TcdC protein from 232 to 61 amino acids. It is possible that different TcdC variants affect toxin production differently, a hypothesis with important implications for the pathogenic potential of variant C. difficile strains.
Figures
FIG. 1.
Primers used in the PaLoc analysis. (A) Specificity and nucleotide sequences of primers and molecular sizes of the PCR products obtained for each pair of primers. (B) Location of PCR primers on a schematic representation of the PaLoc region. The small arrowheads indicate the orientation of primers.
FIG. 2.
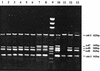
Detection of PaLoc accessory genes _cdd_-3, tcdC, tcdD, tcdE, and _cdu_-2 by multiplex PCR in eight C. difficile strains with variant PaLoc genes identified in this study. Lane 1, Pd 5; lane 2, Pd7; lane 3, Pd13; lane 4, Pd16; lane 5, Pd53; lane 6, Pd 55; lane 7, M7; lane 8, Pd3; lane 9, DNA molecular weight marker IX; lane 10, C. difficile VPI10463; lane 11, ATCC 43597; lane 12, C. difficile 51377; lane 13, C. difficile 57267.
FIG. 3.
Toxinotyping of eight C. difficile strains with variant PaLoc genes and of the reference strains C. difficile VPI 10463, C. difficile 51377, and C. difficile 57267. (For descriptions of the other toxinotypes, see Rupnik et al. [21, 22]). The PCR-RFLP patterns of A3 and B1 fragments are shown for each strain. A3 fragments were digested with _Eco_RI (E), and B1 fragments were digested with _Hin_cII (H) and _Acc_I (A). M, 100-bp DNA ladder (BioLabs). Pd7, Pd13, Pd16, and Pd55 (C. difficile strains), toxinotype V; Pd5 and Pd53, toxinotype VI; Pd3 and M7, toxinotype 0 (C. difficile M7 represents a new toxinotype, as demonstrated by PCR-RFLP analysis of the entire PaLoc). C. difficile VPI 10463, C. difficile 51377, and C. difficile 57267 are the reference strains for toxinotypes 0, VI, and VII, respectively.
FIG. 4.
Comparison of the TcdC nucleotide and amino acid sequences of the C. difficile reference strain VPI 10463 with those of the variant TcdC proteins identified in C. difficile clinical isolates examined in this study. Dots and dashes indicate identical bases and gaps, respectively, for the different tcdC alleles (tcdC-A, -B, and -C). The termination codon in tcdC-A is underlined. Only the amino acid changes are indicated for each TcdC variant. The eight 3-amino-acid repeats of the VPI 10463 TcdC are indicated by open (acidic in nature) and grey (basic in nature) boxes.
Similar articles
- Molecular characterization of pathogenicity locus (PaLoc) and tcdC genetic diversity among tcdA+B+Clostridioides difficile clinical isolates in Tehran, Iran.
Kodori M, Ghalavand Z, Yadegar A, Eslami G, Azimirad M, Krutova M, Abadi A, Zali MR. Kodori M, et al. Anaerobe. 2020 Dec;66:102294. doi: 10.1016/j.anaerobe.2020.102294. Epub 2020 Nov 9. Anaerobe. 2020. PMID: 33181348 - TcdC does not significantly repress toxin expression in Clostridium difficile 630ΔErm.
Bakker D, Smits WK, Kuijper EJ, Corver J. Bakker D, et al. PLoS One. 2012;7(8):e43247. doi: 10.1371/journal.pone.0043247. Epub 2012 Aug 17. PLoS One. 2012. PMID: 22912837 Free PMC article. - Analysis of the pathogenicity locus in Clostridium difficile strains.
Cohen SH, Tang YJ, Silva J Jr. Cohen SH, et al. J Infect Dis. 2000 Feb;181(2):659-63. doi: 10.1086/315248. J Infect Dis. 2000. PMID: 10669352 - Clostridium difficile toxin synthesis is negatively regulated by TcdC.
Dupuy B, Govind R, Antunes A, Matamouros S. Dupuy B, et al. J Med Microbiol. 2008 Jun;57(Pt 6):685-689. doi: 10.1099/jmm.0.47775-0. J Med Microbiol. 2008. PMID: 18480323 Review. - Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes.
Rupnik M. Rupnik M. FEMS Microbiol Rev. 2008 May;32(3):541-55. doi: 10.1111/j.1574-6976.2008.00110.x. Epub 2008 Apr 3. FEMS Microbiol Rev. 2008. PMID: 18397287 Review.
Cited by
- Comparative genomics of zoonotic pathogen Clostridioides difficile of animal origin to understand its diversity.
Karthik K, Anbazhagan S, Priyadharshini MLM, Sharma RK, Manoharan S. Karthik K, et al. 3 Biotech. 2024 Nov;14(11):257. doi: 10.1007/s13205-024-04102-7. Epub 2024 Oct 4. 3 Biotech. 2024. PMID: 39372495 - Pathogenicity and virulence of Clostridioides difficile.
Buddle JE, Fagan RP. Buddle JE, et al. Virulence. 2023 Dec;14(1):2150452. doi: 10.1080/21505594.2022.2150452. Virulence. 2023. PMID: 36419222 Free PMC article. Review. - Characterization of the virulence of three novel clade 2 Clostridioides (Clostridium) difficile strains and a two-year screening in animals and humans in Brazil.
Diniz AN, Moura LNF, Cruz DSG, Oliveira Junior CA, Figueiredo HCP, Cunha JLR, Vilela EG, Kuijper EJ, Wilcox MH, Lobato FCF, Silva ROS. Diniz AN, et al. PLoS One. 2022 Aug 26;17(8):e0273013. doi: 10.1371/journal.pone.0273013. eCollection 2022. PLoS One. 2022. PMID: 36026500 Free PMC article. - Characterization of Clostridioides difficile Strains from an Outbreak Using MALDI-TOF Mass Spectrometry.
Calderaro A, Buttrini M, Farina B, Montecchini S, Martinelli M, Arcangeletti MC, Chezzi C, De Conto F. Calderaro A, et al. Microorganisms. 2022 Jul 21;10(7):1477. doi: 10.3390/microorganisms10071477. Microorganisms. 2022. PMID: 35889196 Free PMC article. - Dogs as carriers of virulent and resistant genotypes of Clostridioides difficile.
Finsterwalder SK, Loncaric I, Cabal A, Szostak MP, Barf LM, Marz M, Allerberger F, Burgener IA, Tichy A, Feßler AT, Schwarz S, Monecke S, Ehricht R, Ruppitsch W, Spergser J, Künzel F. Finsterwalder SK, et al. Zoonoses Public Health. 2022 Sep;69(6):673-681. doi: 10.1111/zph.12956. Epub 2022 May 12. Zoonoses Public Health. 2022. PMID: 35546073 Free PMC article.
References
- Aktories, K., J. Selzer, F. Hofmann, and I. Just. 1997. Molecular mechanisms of action of Clostridium difficile toxins A and B, p. 393-407. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom.
- Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. H. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. - PMC - PubMed
- Bartlett, J. G. 1992. Antibiotic-associated diarrhea. Clin. Infect. Dis. 15: 573-581. - PubMed
- Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases