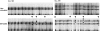Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification - PubMed (original) (raw)
Comparative Study
. 2002 Sep 2;196(5):629-39.
doi: 10.1084/jem.20011693.
Peter McGuire, Hong Zan, Xiao-Jie Yan, Andrea Cerutti, Emilia Albesiano, Steven L Allen, Vincent Vinciguerra, Kanti R Rai, Manlio Ferrarini, Paolo Casali, Nicholas Chiorazzi
Affiliations
- PMID: 12208878
- PMCID: PMC2194006
- DOI: 10.1084/jem.20011693
Comparative Study
Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification
Carmela Gurrieri et al. J Exp Med. 2002.
Abstract
Chronic lymphocytic leukemia (CLL) arises from the clonal expansion of a CD5(+) B lymphocyte that is thought not to undergo intraclonal diversification. Using V(H)DJ(H) cDNA single strand conformation polymorphism analyses, we detected intraclonal mobility variants in 11 of 18 CLL cases. cDNA sequence analyses indicated that these variants represented unique point-mutations (1-35/patient). In nine cases, these mutations were unique to individual submembers of the CLL clone, although in two cases they occurred in a large percentage of the clonal submembers and genealogical trees could be identified. The diversification process responsible for these changes led to single nucleotide changes that favored transitions over transversions, but did not target A nucleotides and did not have the replacement/silent nucleotide change characteristics of antigen-selected B cells. Intraclonal diversification did not correlate with the original mutational load of an individual CLL case in that diversification was as frequent in CLL cells with little or no somatic mutations as in those with considerable mutations. Finally, CLL B cells that did not exhibit intraclonal diversification in vivo could be induced to mutate their V(H)DJ(H) genes in vitro after stimulation. These data indicate that a somatic mutation mechanism remains functional in CLL cells and could play a role in the evolution of the clone.
Figures
Figure 1.
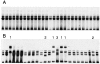
SSCP analysis from two CLL cases displaying either the absence or presence of intraclonal diversification. All 27 VHDJH transcripts from CLL 216 showed identical mobility (A). In CLL 105, seven of the 25 VHDJH transcripts showed a mobility pattern different from that displayed by the remaining 18 VHDJH transcripts (B). Sequence analysis showed that the VHDJH cDNAs 1–3 were all collinear and collinear with the most represented transcripts confirming their monoclonality. However, these transcripts displayed nucleotide variations distributed randomly throughout the VH segment. VHDJH transcripts labeled 1 were identical among themselves but different, though collinear, from the VHDJH transcripts 2 and 3, and the most represented transcripts. VHDJH transcripts 2 were all identical but different, though collinear, from the VHDJH transcripts 1, 3, and the most represented transcripts. VHDJH transcripts 3 were identical but different, though collinear, from the VHDJH transcripts 1, 2 and the most represented transcripts.
Figure 2.
Genealogical tree constructed using VHDJH sequences of CLL nos. 261 and 105. Point-mutations are indicated by their codon number and the nature of the base change. Shared point-mutations and acquired unique point-mutations are indicated above and below the line, respectively. Vertical bars depict S mutations, and lollipops depict R mutations. The putative intermediate elements are depicted with gray nuclei.
Figure 3.
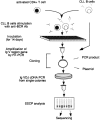
Schematic representation of the steps involved in the in vitro induction of somatic hypermutation in CLL B cells.
Figure 4.
In vitro induction of somatic hypermutation. CLL 136 and CLL 216 did not exhibit evidence of in vivo intraclonal diversity (see Table I). After in vitro stimulation, 3 of 32 VHDJH transcripts in CLL 136 and 5 of 34 in CLL 216 displayed gel mobilities different from that of corresponding transcripts obtained from the unstimulated CLL cells. Transcripts were sequenced and each contained at least one nucleotide change.
Similar articles
- Somatic diversification and selection of immunoglobulin heavy and light chain variable region genes in IgG+ CD5+ chronic lymphocytic leukemia B cells.
Hashimoto S, Dono M, Wakai M, Allen SL, Lichtman SM, Schulman P, Vinciguerra VP, Ferrarini M, Silver J, Chiorazzi N. Hashimoto S, et al. J Exp Med. 1995 Apr 1;181(4):1507-17. doi: 10.1084/jem.181.4.1507. J Exp Med. 1995. PMID: 7535340 Free PMC article. - IgV gene intraclonal diversification and clonal evolution in B-cell chronic lymphocytic leukaemia.
Bagnara D, Callea V, Stelitano C, Morabito F, Fabris S, Neri A, Zanardi S, Ghiotto F, Ciccone E, Grossi CE, Fais F. Bagnara D, et al. Br J Haematol. 2006 Apr;133(1):50-8. doi: 10.1111/j.1365-2141.2005.05974.x. Br J Haematol. 2006. PMID: 16512828 - Molecular analysis of rearranged VH genes during B cell chronic lymphocytic leukemia: intraclonal stability is frequent but not constant.
Korganow AS, Martin T, Weber JC, Lioure B, Lutz P, Knapp AM, Pasquali JL. Korganow AS, et al. Leuk Lymphoma. 1994 Jun;14(1-2):55-69. doi: 10.3109/10428199409049651. Leuk Lymphoma. 1994. PMID: 7920229 - The origin of B-cell chronic lymphocytic leukemia.
Ghia P, Caligaris-Cappio F. Ghia P, et al. Semin Oncol. 2006 Apr;33(2):150-6. doi: 10.1053/j.seminoncol.2006.01.009. Semin Oncol. 2006. PMID: 16616061 Review. - What is the current evidence for antigen involvement in the development of chronic lymphocytic leukemia?
Tobin G, Rosén A, Rosenquist R. Tobin G, et al. Hematol Oncol. 2006 Mar;24(1):7-13. doi: 10.1002/hon.760. Hematol Oncol. 2006. PMID: 16315334 Review.
Cited by
- IgIDivA: immunoglobulin intraclonal diversification analysis.
Zaragoza-Infante L, Junet V, Pechlivanis N, Fragkouli SC, Amprachamian S, Koletsa T, Chatzidimitriou A, Papaioannou M, Stamatopoulos K, Agathangelidis A, Psomopoulos F. Zaragoza-Infante L, et al. Brief Bioinform. 2022 Sep 20;23(5):bbac349. doi: 10.1093/bib/bbac349. Brief Bioinform. 2022. PMID: 36044248 Free PMC article. - Chronic lymphocytic leukemia cells diversify and differentiate in vivo via a nonclassical Th1-dependent, Bcl-6-deficient process.
Patten PE, Ferrer G, Chen SS, Simone R, Marsilio S, Yan XJ, Gitto Z, Yuan C, Kolitz JE, Barrientos J, Allen SL, Rai KR, MacCarthy T, Chu CC, Chiorazzi N. Patten PE, et al. JCI Insight. 2016 Apr 7;1(4):e86288. doi: 10.1172/jci.insight.86288. JCI Insight. 2016. PMID: 27158669 Free PMC article. - Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia.
Gawad C, Pepin F, Carlton VE, Klinger M, Logan AC, Miklos DB, Faham M, Dahl G, Lacayo N. Gawad C, et al. Blood. 2012 Nov 22;120(22):4407-17. doi: 10.1182/blood-2012-05-429811. Epub 2012 Aug 28. Blood. 2012. PMID: 22932801 Free PMC article. - A novel human B cell subpopulation representing the initial germinal center population to express AID.
Kolar GR, Mehta D, Pelayo R, Capra JD. Kolar GR, et al. Blood. 2007 Mar 15;109(6):2545-52. doi: 10.1182/blood-2006-07-037150. Epub 2006 Nov 28. Blood. 2007. PMID: 17132718 Free PMC article. - Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities.
Chiorazzi N, Ferrarini M. Chiorazzi N, et al. Blood. 2011 Feb 10;117(6):1781-91. doi: 10.1182/blood-2010-07-155663. Epub 2010 Dec 9. Blood. 2011. PMID: 21148333 Free PMC article. Review.
References
- Rai, K., and D. Patel. 1995. Chronic lymphocytic leukemia. Hematology: Basic Principles and Practice. 2nd edition. R. Hoffman, E. Benz, S. Shattil, B. Furie, H. Cohen, and L. Silberstein, editors. Churchill Livingstone, New York. pp. 1308–1321.
- Kipps, T.J. 1989. The CD5 B cell. Adv. Immunol. 47:117–185. - PubMed
- Meeker, T.C., J.C. Grimaldi, R. O'Rourke, J. Loeb, G. Juliusson, and S. Einhorn. 1988. Lack of detectable somatic hypermutation in the V region of the Ig H chain gene of a human chronic B lymphocytic leukemia. J. Immunol. 141:3994–3998. - PubMed
- Pratt, L.F., L. Rassenti, J. Larrick, B. Robbins, P.M. Banks, and T.J. Kipps. 1989. Ig V region gene expression in small lymphocytic lymphoma with little or no somatic hypermutation. J. Immunol. 143:699–705. - PubMed
- Kuppers, R., A. Gause, and K. Rajewsky. 1991. B cells of chronic lymphatic leukemia express V genes in unmutated form. Leuk. Res. 15:487–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AR40908/AR/NIAMS NIH HHS/United States
- CA87956/CA/NCI NIH HHS/United States
- AI10811/AI/NIAID NIH HHS/United States
- R01 CA087956/CA/NCI NIH HHS/United States
- R01 AR040908/AR/NIAMS NIH HHS/United States
- AG13910/AG/NIA NIH HHS/United States
- AI07621/AI/NIAID NIH HHS/United States
- R01 AR047872/AR/NIAMS NIH HHS/United States
- R01 CA081554/CA/NCI NIH HHS/United States
- CA81554/CA/NCI NIH HHS/United States
- T32 AI007621/AI/NIAID NIH HHS/United States
- R56 AI045011/AI/NIAID NIH HHS/United States
- AI45011/AI/NIAID NIH HHS/United States
- R01 AI045011/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources