Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants - PubMed (original) (raw)
Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants
Anna N Moraitis et al. Mol Cell Biol. 2002 Oct.
Abstract
Transcriptional regulation by nuclear receptors is controlled by the concerted action of coactivator and corepressor proteins. The product of the thyroid hormone-regulated mammalian gene hairless (Hr) was recently shown to function as a thyroid hormone receptor corepressor. Here we report that Hr acts as a potent repressor of transcriptional activation by RORalpha, an orphan nuclear receptor essential for cerebellar development. In contrast to other corepressor-nuclear receptor interactions, Hr binding to RORalpha is mediated by two LXXLL-containing motifs, a mechanism associated with coactivator interaction. Mutagenesis of conserved amino acids in the ligand binding domain indicates that RORalpha activity is ligand-dependent, suggesting that corepressor activity is maintained in the presence of ligand. Despite similar recognition helices shared with coactivators, Hr does not compete for the same molecular determinants at the surface of the RORalpha ligand binding domain, indicating that Hr-mediated repression is not simply through displacement of coactivators. Remarkably, the specificity of Hr corepressor action can be transferred to a retinoic acid receptor by exchanging the activation function 2 (AF-2) helix. Repression of the chimeric receptor is observed in the presence of retinoic acid, demonstrating that in this context, Hr is indeed a ligand-oblivious nuclear receptor corepressor. These results suggest a novel molecular mechanism for corepressor action and demonstrate that the AF-2 helix can play a dynamic role in controlling corepressor as well as coactivator interactions. The interaction of Hr with RORalpha provides direct evidence for the convergence of thyroid hormone and RORalpha-mediated pathways in cerebellar development.
Figures
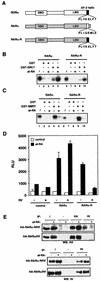
FIG.1.
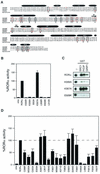
RORα shares common structural and functional determinants with classic nuclear receptors. (A) Primary sequence of RORβ, RORα, and RARγ ligand binding domains. Amino acids involved in the LBP identified by crystallographic analysis are highlighted in red. Amino acids essential for AF-2 activity and known to participate in ligand binding targeted for site-directed mutagenesis are circled and boxed, respectively. The respective amino acid change is indicated below the sequence. The secondary structure is represented by black bars for the α-helices and arrows for the β-sheets. (B) RORα hydrophobic cleft mutants (V335R, K339A, and I353A) and AF-2 helix mutants (L506R, E509K, and L510A) are transcriptionally inactive in transfected Cos-1 cells, with the exception of the cleft mutant K357A. Normalized values are calculated in terms of percent RORα activity with respect to wild type. These results are the average of three independent experiments. (C) Binding of RORα and hydrophobic cleft (K339A, K357A) and AF-2 helix (E509K) mutants to SRC proteins. GST-SRC1aRID, GST-p/CIPRID, and GST-GRIP1RID fusion proteins were coupled to Sepharose beads incubated with 35S-labeled RORα, RORαK339A, RORαK357A, and RORαE509K. The input lane (i) represents 10% of total lysate included in the binding reaction. (D) Cos-1 cells were cotransfected with RORα LBP mutants and ROREα23-TkLuc reporter. Normalized luciferase values are expressed as percent activity with respect to wild-type RORα. These results are the average of three independent experiments.
FIG. 2.
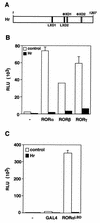
Hr represses ROR transcriptional activation. (A) Schematic representation of the Hr protein containing two LXXLL motifs (LXD1 and LXD2) and two ΦXXΦΦ motifs (ΦxD1 and ΦxD2). The numbers above indicate amino acid positions. (B) Hr represses RORα, -β, and -γ constitutive transcriptional activities. Cos-1 cells were cotransfected with hRORα, mRORβ, mRORγ, and ROREα23-TKLuc in the absence (open bars) or the presence (black bars) of Hr. (C) Hr represses RORα activity on a heterologous promoter through its LBD. Schematic representation of the Gal4-RORαLBD. Numbers above indicate the amino acid positions. Cos-1 cells were cotransfected with Gal4-RORαLBD, Hr, and UAS2TKLuc. Normalized values are presented in relative luciferase units (RLU). A representative experiment of three independent experiments is shown. Error bars represent the standard deviation between duplicate samples.
FIG. 3.
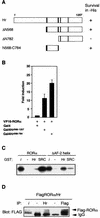
Determinants involved in Hr-RORα interaction. (A) A domain of Hr encoding two LXXLL motifs is sufficient for interaction with RORα. Results of yeast two-hybrid assay with Hr deletion derivatives. The indicated Hr fragments were expressed as fusion proteins with the LexA DBD and tested for interaction with the RORα LBD fused with the VP16 activation domain. +, survival in the absence of histidine. (B) Cos-1 cells were cotransfected with Gal4-Hr568-1207, Gal4-Hr568-784, VP16-RORα, and UAS2TKLuc. Normalized values are presented. (C) The AF-2 helix inhibits Hr binding to RORα in vitro. In vitro-translated and labeled RORα and RORαΔAF-2 were assayed for interaction with GST-SRC1RID or GST-Hr568-784 coupled to Sepharose beads. The input lane (i) represents 10% of total lysate included in each binding reaction. (D) Hr interacts with RORα in vivo. Cos-1 cells were transiently transfected with pCMX-FlagRORα and pRk5-mycHr. Cell lysates were subjected to immunoprecipitation (IP) with Hr antibody, Flag antibody, or rabbit or mouse immunoglobulin G (as negative controls), followed by immunoblotting with anti-Flag. The input lane (i) represents 20% of lysate used in each IP.
FIG. 4.
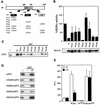
Hr repression requires intact LXXLL motifs. (A) Schematic representation of the Hr protein. Hrm1-Hr-m8 encoding point mutations of the LXD1, LXD2, and ΦXD1 motifs are represented. (B) Hr and Hrm1-Hrm8 expression plasmids were cotransfected into Cos-1 cells with RORα and ROREα23-TkLuc reporter, as shown at the top of the panel. Normalized values are expressed as a percentage of RORα activity. Results are the average of three independent experiments. Cos-1 cells were transiently transfected with pRK5-mycHr wild-type and mutant expression vectors, as shown at the bottom of the panel. Extracts were immunoblotted with Hr antibody. (C) Hr repression correlates with RORα binding. GST-Hr and GST-Hrm1-Hrm8 were coupled to Sepharose beads and incubated with 35S-labeled RORαΔAF2 mutant, in a GST pull-down assay. The input lane (i) represents 10% of total lysate included in each binding reaction. (D) Hr interaction is not mediated through residues of the hydrophobic cleft. 35S-labeled hydrophobic cleft mutants (V335R, K339A, I353A, K357A)/ΔAF2 were assayed for interaction with GST-Hr in a pull-down assay as above. (E) HrRID does not compete with endogenous coactivators. Cos-1 cells were transiently transfected with RORα, Hr and HrRID expression plasmids. Normalized values are expressed as relative luciferase units (RLU). Error bars represent the standard deviation between duplicate samples. This is one representative experiment of three.
FIG.5.
RORα AF-2 helix dictates specificity of Hr repression function. (A) Schematic representation of RORα and RARα, whose AF-2 helix is represented by a solid and an open box, respectively. RARα-R is a chimeric RARα encoding the RORα AF-2 helix. For GST pull-down assays, 35S-labeled RARα and RARα-R were incubated with GST, GST-SRC1RID (B), or GST-SMRTRID (C) in the absence (ethanol) or the presence of 10−6M at-RA. Input (i) represents 10% of the labeled protein used in a binding reaction. (D) Cos-1 cells were cotransfected with TREp3-TkLuc, pCMX (control), hRARα/hRXRα (RARα), or hRARα-R/hRXRα (RARα-R) in the absence (−) or the presence (+) of Hr. Cells were treated with ethanol (open bars) or with 10−8 M at-RA (closed bars). Normalized values are expressed in relative luciferase units (RLU). Error bars represent the standard deviation between duplicate samples. This is a representative experiment of a total of three independent experiments. (E) Hr interacts with RARα-R. Cos-1 cells were transiently transfected with pRK5-myc-rhr, pCMX-HA-RARα-R, or pCMX-HA-RARα. Cells were treated with ethanol (−) or 10−8 M at-RA (+). Cell lysates were subjected to immunoprecipitation (IP) with HA antibody, Hr antibody, or rabbit immunoglobulin G (as negative control), followed by immunoblotting with anti-HA or anti-Hr. The input lanes (i) represents 40% of lysate used in each IP.
Similar articles
- The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes.
Jetten AM, Kurebayashi S, Ueda E. Jetten AM, et al. Prog Nucleic Acid Res Mol Biol. 2001;69:205-47. doi: 10.1016/s0079-6603(01)69048-2. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550795 Review. - Characterization of the retinoid orphan-related receptor-alpha coactivator binding interface: a structural basis for ligand-independent transcription.
Harris JM, Lau P, Chen SL, Muscat GE. Harris JM, et al. Mol Endocrinol. 2002 May;16(5):998-1012. doi: 10.1210/mend.16.5.0837. Mol Endocrinol. 2002. PMID: 11981035 - Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling.
Hsieh JC, Sisk JM, Jurutka PW, Haussler CA, Slater SA, Haussler MR, Thompson CC. Hsieh JC, et al. J Biol Chem. 2003 Oct 3;278(40):38665-74. doi: 10.1074/jbc.M304886200. Epub 2003 Jul 7. J Biol Chem. 2003. PMID: 12847098 - The co-repressor hairless protects RORalpha orphan nuclear receptor from proteasome-mediated degradation.
Moraitis AN, Giguère V. Moraitis AN, et al. J Biol Chem. 2003 Dec 26;278(52):52511-8. doi: 10.1074/jbc.M308152200. Epub 2003 Oct 21. J Biol Chem. 2003. PMID: 14570920 - Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism.
Jetten AM. Jetten AM. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. Epub 2009 Apr 3. Nucl Recept Signal. 2009. PMID: 19381306 Free PMC article. Review.
Cited by
- Gene expression profile of the skin in the 'hairpoor' (HrHp) mice by microarray analysis.
Kim BK, Baek IC, Lee HY, Kim JK, Song HH, Yoon SK. Kim BK, et al. BMC Genomics. 2010 Nov 18;11:640. doi: 10.1186/1471-2164-11-640. BMC Genomics. 2010. PMID: 21083932 Free PMC article. - An overview of nuclear receptor coregulators involved in cerebellar development.
Nishihara E. Nishihara E. Cerebellum. 2008;7(1):48-59. doi: 10.1007/s12311-008-0018-z. Cerebellum. 2008. PMID: 18418685 Review. - Compound heterozygous mutations in the vitamin D receptor in a patient with hereditary 1,25-dihydroxyvitamin D-resistant rickets with alopecia.
Zhou Y, Wang J, Malloy PJ, Dolezel Z, Feldman D. Zhou Y, et al. J Bone Miner Res. 2009 Apr;24(4):643-51. doi: 10.1359/jbmr.081216. J Bone Miner Res. 2009. PMID: 19049339 Free PMC article. - The hairless mouse in skin research.
Benavides F, Oberyszyn TM, VanBuskirk AM, Reeve VE, Kusewitt DF. Benavides F, et al. J Dermatol Sci. 2009 Jan;53(1):10-8. doi: 10.1016/j.jdermsci.2008.08.012. Epub 2008 Oct 19. J Dermatol Sci. 2009. PMID: 18938063 Free PMC article. Review. - Thyroid hormone and cerebellar development.
Anderson GW. Anderson GW. Cerebellum. 2008;7(1):60-74. doi: 10.1007/s12311-008-0021-4. Cerebellum. 2008. PMID: 18418681 Review.
References
- Ahmad, W., M. Faiyaz ul Haque, V. Brancolini, H. C. Tsou, S. ul Haque, H. Lam, V. M. Aita, J. Owen, M. deBlaquiere, J. Frank, P. B. Cserhalmi-Friedman, A. Leask, J. A. McGrath, M. Peacocke, M. Ahmad, J. Ott, and A. M. Christiano. 1998. Alopecia universalis associated with a mutation in the human hairless gene. Science 279:720-724. - PubMed
- Atkins, G. B., X. Hu, M. G. Guenther, C. Rachez, L. P. Freedman, and M. A. Lazar. 1999. Coactivators for the orphan nuclear receptor RORα. Mol. Endocrinol. 13:1550-1557. - PubMed
- Chang, C.-Y., J. D. Norris, H. Grøn, L. A. Paige, P. T. Hamilton, D. J. Kenan, D. Fowlkes, and D. P. McDonnell. 1999. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol. Cell. Biol. 19:8226-8239. - PMC - PubMed
- Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. - PubMed
- Chomez, P., I. Neveu, A. Mansen, E. Kiesler, L. Larsson, B. Vennstrom, and E. Arenas. 2000. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(α) orphan receptor. Development 127:1489-1498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases