Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains - PubMed (original) (raw)
Comparative Study
. 2002 Oct;184(19):5479-90.
doi: 10.1128/JB.184.19.5479-5490.2002.
D Alland, J A Eisen, L Carpenter, O White, J Peterson, R DeBoy, R Dodson, M Gwinn, D Haft, E Hickey, J F Kolonay, W C Nelson, L A Umayam, M Ermolaeva, S L Salzberg, A Delcher, T Utterback, J Weidman, H Khouri, J Gill, A Mikula, W Bishai, W R Jacobs Jr, J C Venter, C M Fraser
Affiliations
- PMID: 12218036
- PMCID: PMC135346
- DOI: 10.1128/JB.184.19.5479-5490.2002
Comparative Study
Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains
R D Fleischmann et al. J Bacteriol. 2002 Oct.
Abstract
Virulence and immunity are poorly understood in Mycobacterium tuberculosis. We sequenced the complete genome of the M. tuberculosis clinical strain CDC1551 and performed a whole-genome comparison with the laboratory strain H37Rv in order to identify polymorphic sequences with potential relevance to disease pathogenesis, immunity, and evolution. We found large-sequence and single-nucleotide polymorphisms in numerous genes. Polymorphic loci included a phospholipase C, a membrane lipoprotein, members of an adenylate cyclase gene family, and members of the PE/PPE gene family, some of which have been implicated in virulence or the host immune response. Several gene families, including the PE/PPE gene family, also had significantly higher synonymous and nonsynonymous substitution frequencies compared to the genome as a whole. We tested a large sample of M. tuberculosis clinical isolates for a subset of the large-sequence and single-nucleotide polymorphisms and found widespread genetic variability at many of these loci. We performed phylogenetic and epidemiological analysis to investigate the evolutionary relationships among isolates and the origins of specific polymorphic loci. A number of these polymorphisms appear to have occurred multiple times as independent events, suggesting that these changes may be under selective pressure. Together, these results demonstrate that polymorphisms among M. tuberculosis strains are more extensive than initially anticipated, and genetic variation may have an important role in disease pathogenesis and immunity.
Figures
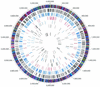
FIG.1.
Circular representation of the M. tuberculosis chromosome illustrating the location of each predicted protein-coding region as well as selected features differing between the CDC1551 and H37Rv strains. The outer concentric circle shows predicted protein-coding regions on both strands, color coded according to role category. The second concentric circle shows the location of nonsynonymous substitutions (black). The third concentric circle shows the location of synonymous substitutions (blue). The fourth concentric circle shows the location of substitutions in noncoding regions (red). The fifth concentric circle shows the location of insertions in strain CDC1551, including coding (black) and noncoding (blue) regions, and the location of phage phiRv1 (red). The sixth concentric circle shows the location of insertions in strain H37Rv, including coding (black) and noncoding (blue) regions, and the location of phage phiRv1 (red). The seventh concentric circle shows the location of IS_6110_ insertion elements in strains CDC1551 (blue) and H37Rv (red). The eighth (innermost) concentric circle shows the location of tRNAs (blue) and rRNA (red).
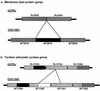
FIG. 2.
(a) Schematic diagram of homologous genome region in strains H37Rv and CDC1551 encoding several membrane lipoproteins. The region in strain H37Rv contains two genes in tandem (Rv2543 and Rv2544) that are 87% identical to each other at the protein level. The homologous region in strain CDC1551 contains three genes (MT2618, MT2619, and MT2620), with the middle gene, MT2619, being unique to strain CDC1551 and 88 and 84% identical to MT2619 and MT2620, respectively. Homology between strain CDC1551 and M. bovis and equivalent evolutionary distances between paralogs suggest that the three paralogs arose in a common ancestor of the M. tuberculosis complex and subsequent loss of MT2619 occurred in the H37Rv lineage. (b) Schematic diagram of the tandem adenylate cyclase region. Two paralogous cyclases flank the region (MT1359/Rv1318c and MT1362/Rv1320c). Analysis revealed two cyclases (MT1360 and MT1361) between the two flanking genes in strain CDC1551 and only one cyclase (Rv1319c) in strain H37Rv. Rv1319c appears to be a chimera of the 5′ half of MT1361 and the 3′ half of MT1360. The 3′ halves of all orthologs share >80% nucleotide identity, while the 5′ halves appear diverse. Phylogenetic analysis indicates that the duplication events share a similar evolutionary distance. Inspection of the M. bovis sequence data reveals that this region is organized in an identical way to the H37Rv genome.
FIG. 3.
The distribution of CDC1551/H37Rv LSPs in clinical M. tuberculosis strains using a slot blot cross hybridization method. Strains include unique isolates and clustered strains with and without epidemiological links. Clusters are grouped by letter (and number if subtyped by a secondary fingerprinting method). Strains with epidemiological links are designated by links. Probes to LSPs are described in Table 3.
FIG. 4.
M. tuberculosis strain phylogeny based on a combination of LSPs, SNPs, and selected phenotypic traits. The tree shown is a consensus of the most parsimonious trees found using the heuristic search algorithm. Character state boxes are shown at the top (for LSPs, a blue box indicates presence and a yellow box indicates absence; for SNPs and other characters, colors correspond to different character states). Characters 1 to 12 are SNPs, characters 13 to 28 are LSPs, character 29 belongs to the Musser group, character 30 is Smear positive or negative, and character 31 is Site (pulmonary or extrapulmonary). Unresolved branching patterns are collapsed.
FIG. 5.
Consistency index for characters in the phylogenetic tree based on the LSPs and SNPs. Each column corresponds to the consistency index for a particular character (SNPs and LSPs). Each bar shows the minimum (black), average (striped), and maximum (grey) value of the consistency index over a large number of different trees (including multiple equally parsimonious trees and multiple distance trees). The percent variability indicates the variability of each marker in the 28 isolates tested. Calculations of the consistency indices were made using the MacClade program.
Similar articles
- Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints.
McEvoy CR, Cloete R, Müller B, Schürch AC, van Helden PD, Gagneux S, Warren RM, Gey van Pittius NC. McEvoy CR, et al. PLoS One. 2012;7(4):e30593. doi: 10.1371/journal.pone.0030593. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496726 Free PMC article. - Differential carriage of virulence-associated loci in the New Zealand Rangipo outbreak strain of Mycobacterium tuberculosis.
Gautam SS, Mac Aogáin M, Bower JE, Basu I, O'Toole RF. Gautam SS, et al. Infect Dis (Lond). 2017 Sep;49(9):680-688. doi: 10.1080/23744235.2017.1330553. Epub 2017 May 23. Infect Dis (Lond). 2017. PMID: 28535727 - Limitations of the Mycobacterium tuberculosis reference genome H37Rv in the detection of virulence-related loci.
O'Toole RF, Gautam SS. O'Toole RF, et al. Genomics. 2017 Oct;109(5-6):471-474. doi: 10.1016/j.ygeno.2017.07.004. Epub 2017 Jul 22. Genomics. 2017. PMID: 28743540 Review. - Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv.
Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. Zheng H, et al. PLoS One. 2008 Jun 11;3(6):e2375. doi: 10.1371/journal.pone.0002375. PLoS One. 2008. PMID: 18584054 Free PMC article. - PE and PPE Genes: A Tale of Conservation and Diversity.
Delogu G, Brennan MJ, Manganelli R. Delogu G, et al. Adv Exp Med Biol. 2017;1019:191-207. doi: 10.1007/978-3-319-64371-7_10. Adv Exp Med Biol. 2017. PMID: 29116636 Review.
Cited by
- Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints.
McEvoy CR, Cloete R, Müller B, Schürch AC, van Helden PD, Gagneux S, Warren RM, Gey van Pittius NC. McEvoy CR, et al. PLoS One. 2012;7(4):e30593. doi: 10.1371/journal.pone.0030593. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496726 Free PMC article. - Aminoglycoside multiacetylating activity of the enhanced intracellular survival protein from Mycobacterium smegmatis and its inhibition.
Chen W, Green KD, Tsodikov OV, Garneau-Tsodikova S. Chen W, et al. Biochemistry. 2012 Jun 19;51(24):4959-67. doi: 10.1021/bi3004473. Epub 2012 Jun 5. Biochemistry. 2012. PMID: 22646013 Free PMC article. - Mycobacterium tuberculosis genome-wide screen exposes multiple CD8 T cell epitopes.
Hammond AS, Klein MR, Corrah T, Fox A, Jaye A, McAdam KP, Brookes RH. Hammond AS, et al. Clin Exp Immunol. 2005 Apr;140(1):109-16. doi: 10.1111/j.1365-2249.2005.02751.x. Clin Exp Immunol. 2005. PMID: 15762882 Free PMC article. - Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions.
Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Gey van Pittius NC, et al. BMC Evol Biol. 2006 Nov 15;6:95. doi: 10.1186/1471-2148-6-95. BMC Evol Biol. 2006. PMID: 17105670 Free PMC article. - The antigenome: from protein subunit vaccines to antibody treatments of bacterial infections?
Giefing C, Nagy E, von Gabain A. Giefing C, et al. Adv Exp Med Biol. 2009;655:90-117. doi: 10.1007/978-1-4419-1132-2_9. Adv Exp Med Biol. 2009. PMID: 20047038 Free PMC article. Review.
References
- Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. - PubMed
- Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. - PubMed
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Beck-Sague, C. S., W. Dooley, M. D. Hutton, J. Otten, A. Breeden, J. T. Crawford, A. E. Pitchenik, C. Woodley, G. Cauthen, and W. R. Jarvis. 1992. Hospital outbreak of multidrug-resistant Mycobacterium tuberculosis infections. Factors in transmission to staff and HIV-infected patients. JAMA 268:1280-1286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases