Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest - PubMed (original) (raw)
Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest
Rebecca L Read et al. Proc Natl Acad Sci U S A. 2002.
Abstract
The S-M checkpoint delays mitosis until DNA replication is complete; cells defective in this checkpoint lose viability when DNA replication is inhibited. This inviability can be suppressed in fission yeast by overexpression of Cid1 or the related protein Cid13. Fission yeast contain six cid1/cid13-like genes, whereas budding yeast has just two, TRF4 and TRF5. Trf4 and Trf5 were recently reported to comprise an essential DNA polymerase activity required for the establishment of sister chromatid cohesion. In contrast, we find that Cid1 is not a DNA polymerase but instead uses RNA substrates and has poly(A) polymerase activity. Unlike the previously characterized yeast poly(A) polymerase, which is a nuclear enzyme, Cid1 and Cid13 are constitutively cytoplasmic. Cid1 has a degree of substrate specificity in vitro, consistent with the notion that it targets a subset of cytoplasmic mRNAs for polyadenylation in vivo, hence increasing their stability and/or efficiency of translation. Preferred Cid1 targets presumably include mRNAs encoding components of the S-M checkpoint, whereas Cid13 targets are likely to be involved in dNTP metabolism. Cytoplasmic polyadenylation is known to be an important regulatory mechanism during early development in animals. Our findings in yeast suggest that this level of gene regulation is of more general significance in eukaryotic cells.
Figures
Fig 1.
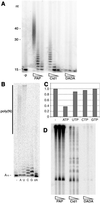
Recombinant Cid1 is an ATPase but lacks Pol activity. (A) Purified recombinant Cid1 and Cid1DADA were separated by SDS/PAGE and silver-stained. The sizes of the molecular mass markers in the lane next to Cid1 are indicated on the left. (B) ATPase activities (μmol min−1) in the presence of no added protein or 250 ng of recombinant Cid1 or Cid1DADA, as indicated; the average of three independent determinations is shown graphically (error bars represent one standard deviation). (C) In vitro Pol assays were performed by using a dT25/dA40 substrate and 5-fold serial dilutions of E. coli PolI large fragment (Klenow), Cid1, or Cid1DADA as indicated. Control reactions included either no added protein (−p) or Klenow enzyme in the absence of added dNTPs (−d). The sizes of reaction products in nucleotides are indicated on the left.
Fig 2.
Cid1 is a poly(A) polymerase. (A) In vitro poly(A) polymerase assays were performed by using 50, 5, and 0.5 ng of S. cerevisiae poly(A) polymerase (PAP), and 200, 40, and 8 ng of Cid1 or Cid1DADA, as indicated, and a radiolabeled A15 RNA substrate for 10 min at 30°C. A control reaction included no added protein (−p). The sizes of reaction products in nucleotides are indicated on the left. (B) Poly(A) polymerase assays were performed for 30 min at 30°C by using 400 ng of Cid1 and ATP (A), UTP (U), CTP (C), GTP (G), dATP (dA), or no added nucleotide triphosphate (−). (C) Cid1 ATPase assays were performed as in Fig. 1_B_ (−) or with the addition of a 5-fold molar excess of each of the unlabeled NTPs indicated. (D) Poly(A) polymerase assays were performed as in A, but using 2-fold serial dilutions of each enzyme, total S. pombe RNA, and [α-32P]cordycepin 5′-triphosphate.
Fig 3.
Cid13 is a Cid1- and poly(A) polymerase-related protein that suppresses the HU sensitivity of rad3 mutants. (A) Alignment of amino acid sequences of the putative catalytic cores of Cid1, Cid11, Cid13, and S. pombe poly(A) polymerase. Amino acid residue numbers are indicated to the left of each sequence, conserved residues are highlighted on a black background, and conservative substitutions are shaded. The Pol β superfamily motif is underlined, and the two conserved aspartate residues changed to alanine in the Cid1 DADA mutant are indicated by asterisks. (B and C) Survival of _rad3_ts or _rad3_kd cells transformed with pUR-cid13 (_cid13_oe) or pUR (vector), as indicated, after plating onto agar containing various doses of HU (B) or after UV irradiation on agar plates (C). Survival was scored by counting colonies after 4 days of incubation at 32°C. (D) Survival of the same strains after short-term exposure to 5 mM HU in liquid culture at 32.5°C. Aliquots of cells taken at the times indicated were scored for viability by plating onto agar lacking HU and incubation at 32°C for 3 days. (E) Cells of the _rad3_ts strain transformed with pUR-cid13 (_cid13_oe) or pUR (vector), as indicated, were exposed to 5 mM HU at 32°C in duplicate liquid cultures for 3 h and were then assayed for Cds1 kinase activity in vitro using myelin basic protein (MBP) as substrate. (F) The strains indicated were inoculated onto plates containing various doses of HU, and survival was scored by counting colonies after 4–5 days of incubation at 32°C.
Fig 4.
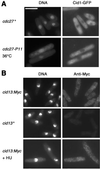
Cid1 and Cid13 are cytoplasmic proteins. (A) Fluorescence micrographs of living cid1-GFP cells stained briefly with Hoechst 33258 to reveal DNA (Left) and GFP fluorescence (Right). (Upper) cid1-GFP (cdc27+) cells grown at 32°C. (Lower) cid1-GFP cdc27-P11 cells shifted to 36°C for 4 h. (B) Micrographs of fixed wild-type (cid13+; negative control) and cid13:Myc cells, as indicated, processed for anti-Myc immunofluorescence (Right) and stained with DAPI to reveal DNA (Left). The cells in the lower panels were exposed to 20 mM HU for 3 h before fixation. (Scale bar: 10 μm.)
Fig 5.
Expression of Suc22 does not depend on Cid1 or Cid13. (A) Suc22 protein levels in whole-cell lysates from wild-type (w.t.), _cid1_Δ, and _cid13_Δ strains were estimated by immunoblotting with a polyclonal anti-Suc22 antibody (Upper Left). Protein loading was controlled by reprobing the same filter with an anti-Cdc2 antibody. Suc22 levels in two independent experiments were quantified by densitometry and expressed graphically (Right) after normalizing for Cdc2 levels. (B) Suc22 and Cdc2 immunoblotting was performed as in A from parallel cultures that had been exposed (+) or not (−) to 10 mM HU for 4 h at 30°C.
Similar articles
- Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated.
Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. Wang SW, et al. Mol Cell Biol. 2000 May;20(9):3234-44. doi: 10.1128/MCB.20.9.3234-3244.2000. Mol Cell Biol. 2000. PMID: 10757807 Free PMC article. - Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA.
Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P. Saitoh S, et al. Cell. 2002 May 31;109(5):563-73. doi: 10.1016/s0092-8674(02)00753-5. Cell. 2002. PMID: 12062100 - Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control.
Win TZ, Stevenson AL, Wang SW. Win TZ, et al. Mol Cell Biol. 2006 Jun;26(12):4435-47. doi: 10.1128/MCB.02205-05. Mol Cell Biol. 2006. PMID: 16738311 Free PMC article. - The Cid1 family of non-canonical poly(A) polymerases.
Stevenson AL, Norbury CJ. Stevenson AL, et al. Yeast. 2006 Oct 15;23(13):991-1000. doi: 10.1002/yea.1408. Yeast. 2006. PMID: 17072891 Review. - The Cid1 poly(U) polymerase.
Rissland OS, Norbury CJ. Rissland OS, et al. Biochim Biophys Acta. 2008 Apr;1779(4):286-94. doi: 10.1016/j.bbagrm.2008.03.003. Epub 2008 Mar 19. Biochim Biophys Acta. 2008. PMID: 18371314 Review.
Cited by
- Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity.
Lunde BM, Magler I, Meinhart A. Lunde BM, et al. Nucleic Acids Res. 2012 Oct;40(19):9815-24. doi: 10.1093/nar/gks740. Epub 2012 Aug 9. Nucleic Acids Res. 2012. PMID: 22885303 Free PMC article. - Trypanosome MTR4 is involved in rRNA processing.
Cristodero M, Clayton CE. Cristodero M, et al. Nucleic Acids Res. 2007;35(20):7023-30. doi: 10.1093/nar/gkm736. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940093 Free PMC article. - Terminal RNA uridylyltransferases of trypanosomes.
Aphasizhev R, Aphasizheva I. Aphasizhev R, et al. Biochim Biophys Acta. 2008 Apr;1779(4):270-80. doi: 10.1016/j.bbagrm.2007.12.007. Epub 2007 Dec 23. Biochim Biophys Acta. 2008. PMID: 18191648 Free PMC article. - Identification and characterization of nuclear non-canonical poly(A) polymerases from Trypanosoma brucei.
Etheridge RD, Clemens DM, Gershon PD, Aphasizhev R. Etheridge RD, et al. Mol Biochem Parasitol. 2009 Mar;164(1):66-73. doi: 10.1016/j.molbiopara.2008.11.004. Epub 2008 Nov 25. Mol Biochem Parasitol. 2009. PMID: 19070634 Free PMC article. - Terminal uridyltransferase enzyme Zcchc11 promotes cell proliferation independent of its uridyltransferase activity.
Blahna MT, Jones MR, Quinton LJ, Matsuura KY, Mizgerd JP. Blahna MT, et al. J Biol Chem. 2011 Dec 9;286(49):42381-42389. doi: 10.1074/jbc.M111.259689. Epub 2011 Oct 17. J Biol Chem. 2011. PMID: 22006926 Free PMC article.
References
- Melo J. & Toczyski, D. (2002) Curr. Opin. Cell Biol. 14, 237-245. - PubMed
- Uhlmann F. (2001) Curr. Opin. Cell Biol. 13, 754-761. - PubMed
- Wang Z., Castano, I. B., De Las Penas, A., Adams, C. & Christman, M. F. (2000) Science 289, 774-779. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases