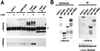Distinct chromosome segregation roles for spindle checkpoint proteins - PubMed (original) (raw)
Distinct chromosome segregation roles for spindle checkpoint proteins
Cheryl D Warren et al. Mol Biol Cell. 2002 Sep.
Abstract
The spindle checkpoint plays a central role in the fidelity of chromosome transmission by ensuring that anaphase is initiated only after kinetochore-microtubule associations of all sister chromatid pairs are complete. In this study, we find that known spindle checkpoint proteins do not contribute equally to chromosome segregation fidelity in Saccharomyces cerevisiae. Loss of Bub1 or Bub3 protein elicits the largest effect. Analysis of Bub1p reveals the presence of two molecular functions. An N-terminal 608-amino acid (nonkinase) portion of the protein supports robust checkpoint activity, and, as expected, contributes to chromosome segregation. A C-terminal kinase-encoding segment independently contributes to chromosome segregation through an unknown mechanism. Both molecular functions depend on association with Bub3p. A 156-amino acid fragment of Bub1p functions in Bub3p binding and in kinetochore localization by one-hybrid assay. An adjacent segment is required for Mad1p binding, detected by deletion analysis and coimmunoprecipitation. Finally, overexpression of wild-type BUB1 or MAD3 genes leads to chromosome instability. Analysis of this activity indicates that the Bub3p-binding domain of Bub1p contributes to this phenotype through disruption of checkpoint activity as well as through introduction of kinetochore or spindle damage.
Figures
Figure 1
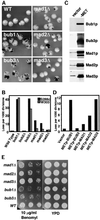
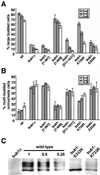
Spindle checkpoint mutants exhibit different rates of chromosome loss. (A) Null mutant sectoring phenotypes. The strains shown are wild type (YPH278), bub1Δ (YFP2), bub3Δ (YFS1100), mad1Δ (YFS1120), mad2Δ (YCD173), and mad3Δ (YFS1205). (B) Chromosome loss rates in null mutants determined by half-sector analysis. Wild type: 49 half-sectored colonies/61,276 total colonies (YPH278); 8/9,305 (YKH231). bub1Δ: 192/4,784 (YFP2); 89/2,121 (YRJ112). bub2Δ: 17/22,101 (YCD165); 3/3,440 (YRJ113). bub3Δ: 137/3,362 (YFS1100); 63/2,237 (YRJ114). mad1Δ: 139/12,394 (YFS1120); 32/8,552 (YMB111). mad2Δ: 87/10,153 (YCD173); 9/3,106 (YMB113). mad3Δ: 57/29,364 (YFS1205); 29/13,186 (YRJ111). (C) Immunoblots showing overexpression from a MET25 promoter. Extracts were taken after 2 h of inductionin media lacking methionine and were analyzed by Western blot using antibody specific for each protein. The left lane (vector, p423MET) shows the endogenous Bub1p expression level where detected; the right lane (–MET) shows protein expressed from the MET25 promoter. All strains were generated from YKH231 by introduction of p423MET-derived plasmids containing full-length open reading frames cloned adjacent to the MET25 promoter. (D) Chromosome loss associated with overexpression of checkpoint genes. Half-sector analysis was performed after plating the strains in C on plates lacking methionine. Vector: 14/19,030. METpBUB1: 195/17,640. METpBUB3: 17/17,875. METpMAD1: 28/11,920. METpMAD2: 92/17,065. METpMAD3: 137/12,115. Two or more additional independent transformants tested for each construct showed the same chromosome instability phenotype by colony sectoring assay. (E) Benomyl sensitivity of checkpoint null mutants. Log phase cultures were spotted in a 10-fold dilution series on rich medium (YPD) or rich medium plus Benomyl. Strains were mad1Δ (YMB111), mad2Δ (YMB113), mad3Δ (YRJ111), bub1Δ (YRJ112), and bub3Δ (YRJ114).
Figure 2
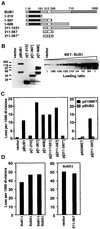
The N terminus of Bub1p contributes to the overexpression phenotype and is counterbalanced by additional BUB3. (A) Diagram of BUB1 protein and protein fragments. The boxes indicate positions of conserved regions of BUB1p. Black: Mad3 like. White: Bub3 binding. Hatched: kinase domain. Star: E333K mutation. (B) Western blot detection of BUB1 overexpression alleles. Left: Wild-type cells (YPH278) containing MET25-promoted Bub1 alleles in p423MET were grown in the absence of methionine. Western blot analysis using an antibody raised to the N-terminal 216 amino acids of Bub1p (Brady and Hardwick, 2000) detects protein bands with migrations consistent with eachconstruct, in addition to faster migrating degradation products. Endogenous Bub1p is not detected at this exposure (vector lane). Right: A dilution series Western blot indicates that the overexpression level for full-length BUB1 is ∼50-fold. (C) Chromosome missegregation induced by BUB1 overexpression alleles. Left: Full-length and partial BUB1 alleles expressed from the MET25 promoter of p423MET were introduced into a wild-type strain (YPH278). Chromosome loss was determined by half-sector analysis after plating to methionine-free medium. Vector (no insert) and pBUB1 data are from D. p[1-210]: 25/14,087. p[1-367]: 127/5,741. p[1-608]: 96/6,592. p[211-1021]: 157/10,697. p[211-367]: 247/13,781. p[211-367*]: 9/5,117. Right: Chromosome loss was determined in YPH278 containing plasmid pairs as shown. p423MET + p415MET: 1/2,616. p423MET + pBUB3: 7/4,998. p[211-367] + p415MET: 37/3,015. p[211-367] + pBUB3: 6/4,510. At least two additional independent transformants of each construct were tested by visual sectoring assay and showed the same chromosome instability phenotype. (D) Chromosome loss in bub1 and bub3 null mutants. Left: Half sector analysis was used to compare chromosome loss rates of bub1Δ, bub3Δ, and bub1Δ bub3Δ mutant strains derived from sporulation of a wild-type diploid (YCD251) into which heterozygous bub1Δ::natMX and bub3Δ::kanMX alleles were introduced by transformation. bub1Δ: 186/5019 (one spore); bub3Δ: 262/5406 (one spore); bub1Δ bub3Δ: 601/12566 (four spores). Right: Chromosome missegration in a bub3Δ strain (YFS1100) containing the vector p423MET (284/5,675) or overexpression plasmid p[211-367] (289/6,069).
Figure 3
Localization of activation domain fusions to kinetochores in a one-hybrid assay. In the one-hybrid assay, fusion of the GAL4-activation domain to a kinetochore-binding protein induces transcription of a centromere-adjacent HIS3 reporter allele (Ortiz et al., 1999). GAL4AD fusion constructs were introduced into strain YJL128. The fusion moiety is indicated to the right; GAL4AD-CTF13 (top) served as a positive control. Four independent transformants were grown to saturation in SD-LEU, diluted to 1.5 × 107 cells/ml, and spotted (3 μl) on SD-HIS, LEU + 5 mM 3-AT. The spots shown were incubated at 30°C for 14 d. The large papillae that appear occasionally in the GAL4AD-BUB1 transformants (observed in ∼25% of transformants) may reflect the occurrence of truncating mutations. All constructs were similarly tested in YJL148, a strain containing a mutant centromere sequence adjacent to the HIS3 reporter (Ortiz et al., 1999). No growth above vector background was observed in these controls. All fusion constructs shown were functional in a two-hybrid assay.
Figure 4
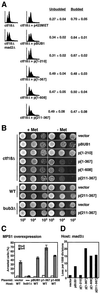
Overexpression of Bub1p or Bub1p fragments both disrupts the spindle checkpoint and causes damage. (A) Disruption of a ctf18Δ-induced checkpoint delay. ctf18Δ strains containing p423MET plasmids expressing no (vector), full-length (BUB1), or partial (1–210, 1–367, 211–367, 1–608) alleles of BUB1 were created by transformation of YFS377. Cells were grown to early log phase in the absence of methionine for 18–24 h, and were prepared for flow cytometry (as in Hanna et al., 2001). A representative histogram is given for each genotype; four independent transformants were analyzed. The fractions of budded and unbudded cells were determined and are shown as mean ± SD (3 d.f.). (B) Disruption of the ctf18Δ delay results in decreased viability. Two independent isolates from each of the ctf18Δ strains described in A were grown to early log phase in media containing methionine. Cells were spotted onto solid medium without methionine in a 10-fold serial dilution series. Overexpression of bub1-[211-367]p in wild-type or bub3Δ cells did not result in a significant reduction in mortality (bottom). (C) Competence to arrest in response to MPS1 overexpression. Log phase cells grown in raffinose media lacking methionine (to induce expression of the BUB1 alleles) were treated with 3% galactose to induce MPS1 expression. Samples were taken at t = 0 and t = 4 h, formaldehyde fixed, DAPI stained, and scored for bud and nuclear morphology. The graph shows the percentage arrested (large-budded uninucleate) cells at each time point. Two independent transformants were analyzed (average ± range indicated). All strains were derived from YML101 (GAL-MPS1). All plasmids overexpressing BUB1 alleles in this strain were derived from p423MET (vector). YCD362 (bub1Δ) was included as a control for the assay. (D) Chromosome missegregation in a mad3Δ host. Chromosome loss rates were determined by half-sector analysis on plates lacking methionine. Vector: 5/3,327. pBUB1: 18/2,295. p[1–210]: 4/2,408. p[1–367]: 51/2,500. p[1–608]: 42/2,766. p[211–367]: 37/2,349. Data for a representative transformant are shown; at least two independent transformants were analyzed for each construct. The strains were YFS1205 derivatives created by introduction of p423MET (vector) and related BUB1 allele overexpression plasmids.
Figure 5
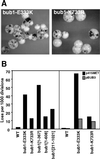
Chromosome missegregation associated with genomic alleles of BUB1. (A) Colony sectoring phenotypes of bub1-E333K (bub1-1) and bub1-K733R. Strains were YCD280 and YCD281. (B) Chromosome loss caused by genomic alleles. Left: Half sector analysis was used to analyze the mutants as shown. WT: 8/8,503. bub1-E333K: 248/5,964. bub1-K733R: 83/6,576. bub1[1-210]: 354/6,769. bub1[1-367]: 477/9,019. bub1[1-608]: 222/14,105. bub1[211-1021]: 355/14,837. Strains were YCD279, YCD280, YCD281, YCD371, YCD358, and YCD407. Right: A centromere plasmid containing a MET25-inducible BUB3 gene (pBUB3) or vector alone (p415MET) was introduced into wild-type (WT, YPH278), bub1-E333K (YCD280), or bub1-K733R (YCD281) strains. Half sector analysis was performed after plating on methionine-free media. WT + p415MET: 1/3,728. WT + pBUB3: 3/4,925. bub1-E333K + p415MET: 104/1,567. bub1-E333K + pBUB3: 48/3,941. bub1-K733R + p415MET: 53/3,654. bub1-K733R + pBUB3: 36/4,102.
Figure 6
Checkpoint competence of BUB1 genomic alleles. (A) Cell cycle arrest. Logarithmically growing cultures of strains containing integrated alleles were transferred to YPD + 15 μg/ml nocodazole. At t = 0, 4, 6, and 8 h after shift into nocodazole, aliquots were formaldehyde fixed and stained with DAPI. Two hundred cells from each were scored for the arrested fraction (large-budded uninucleate cells), and the mean ± standard deviation for three independent integrants is shown. The strains were YPH278, YFP2, YCD371, YCD358, YCD281, and YCD280. (B) Failure of cell cycle arrest. The same samples were scored for the fraction of cells that exhibited multibudded uninucleate cells, an indication mitotic exit in the absence of nuclear division. (C) Anti-Bub1p immunoprecipitates from the genotypes indicated were analyzed by Western blot for the presence and abundance of Bub1 protein (as described in Brady and Hardwick, 2000). Immunoprecipitation product from the wild-type cells was loaded in a dilution series (1, 0.5, and 0.25) for comparison with lanes containing immunoprecipitations from mutant extracts. The strains were YFP2, YPH278, YCD280, and YCD281.
Figure 7
Bub1-[1-608]p associates with Mad1p and Bub3p. (A) Coimmunoprecipitation of full-length Bub1p and bub1-[1-608]p with Mad1p. The strains shown, containing integrated Bub1 alleles expressed from the wild-type BUB1 promoter, were grown to log phase and were incubated with ± 15 μg/ml nocodazole for 2 h at 24°C. Immunoprecipitates were prepared using an α-Bub1p antibody, separated by SDS-PAGE, and transferred to nitrocellulose. The immunoblots were then probed with α-Bub1p and α-Mad1p rabbit antibodies as indicated. The strong band labeled (*) in the Bub1 blot is IgG heavy chain from the immunoprecipitation. Strains shown are YPH278, YFP2, YCD358, and YCD371. (B) Coimmunoprecipitation of full-length Bub1p, bub1-[1-367]p, and bub1-[1-608]p with Bub3p. All strains contained a BUB3-myc allele in the genome. The experimental strains contained a wild-type BUB1 gene in addition to episomal MET25-promoted alleles as indicated. A bub1Δ strain served as control. Left: Bub1p Western blot using a rabbit α-Bub1p antibody. Right: Immunoprecipitation with an α-myc antibody recovered an equivalent amount of myc-tagged Bub3 protein (bottom). The immunoprecipitates were probed with rabbit α-Bub1p antibody (top). The strains were YKH300 (bub1Δ) or YKH238 with pBUB1, p[1-210], p[1-367], or p[1-608].
Similar articles
- The A78V mutation in the Mad3-like domain of Schizosaccharomyces pombe Bub1p perturbs nuclear accumulation and kinetochore targeting of Bub1p, Bub3p, and Mad3p and spindle assembly checkpoint function.
Kadura S, He X, Vanoosthuyse V, Hardwick KG, Sazer S. Kadura S, et al. Mol Biol Cell. 2005 Jan;16(1):385-95. doi: 10.1091/mbc.e04-07-0558. Epub 2004 Nov 3. Mol Biol Cell. 2005. PMID: 15525673 Free PMC article. - Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function.
Brady DM, Hardwick KG. Brady DM, et al. Curr Biol. 2000 Jun 1;10(11):675-8. doi: 10.1016/s0960-9822(00)00515-7. Curr Biol. 2000. PMID: 10837255 - The C-terminal half of Saccharomyces cerevisiae Mad1p mediates spindle checkpoint function, chromosome transmission fidelity and CEN association.
Kastenmayer JP, Lee MS, Hong AL, Spencer FA, Basrai MA. Kastenmayer JP, et al. Genetics. 2005 Jun;170(2):509-17. doi: 10.1534/genetics.105.041426. Epub 2005 Mar 31. Genetics. 2005. PMID: 15802513 Free PMC article. - Regulation of APC-Cdc20 by the spindle checkpoint.
Yu H. Yu H. Curr Opin Cell Biol. 2002 Dec;14(6):706-14. doi: 10.1016/s0955-0674(02)00382-4. Curr Opin Cell Biol. 2002. PMID: 12473343 Review. - Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint.
Elowe S. Elowe S. Mol Cell Biol. 2011 Aug;31(15):3085-93. doi: 10.1128/MCB.05326-11. Epub 2011 May 31. Mol Cell Biol. 2011. PMID: 21628528 Free PMC article. Review.
Cited by
- A dominant interfering Bub1 mutant is insufficient to induce or alter thymic tumorigenesis in vivo, even in a sensitized genetic background.
Cowley DO, Muse GW, Van Dyke T. Cowley DO, et al. Mol Cell Biol. 2005 Sep;25(17):7796-802. doi: 10.1128/MCB.25.17.7796-7802.2005. Mol Cell Biol. 2005. PMID: 16107724 Free PMC article. - Kre29p is a novel nuclear protein involved in DNA repair and mitotic fidelity in Candida glabrata.
Miyazaki T, Tsai HF, Bennett JE. Miyazaki T, et al. Curr Genet. 2006 Jul;50(1):11-22. doi: 10.1007/s00294-006-0072-3. Epub 2006 Apr 28. Curr Genet. 2006. PMID: 16775745 - Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function.
Anderson VE, Prudden J, Prochnik S, Giddings TH Jr, Hardwick KG. Anderson VE, et al. Mol Biol Cell. 2007 Jun;18(6):2047-56. doi: 10.1091/mbc.e06-10-0918. Epub 2007 Mar 28. Mol Biol Cell. 2007. PMID: 17392514 Free PMC article. - The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation.
Taylor SS, Scott MI, Holland AJ. Taylor SS, et al. Chromosome Res. 2004;12(6):599-616. doi: 10.1023/B:CHRO.0000036610.78380.51. Chromosome Res. 2004. PMID: 15289666 Review. - A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint.
Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. Moyle MW, et al. J Cell Biol. 2014 Mar 3;204(5):647-57. doi: 10.1083/jcb.201311015. Epub 2014 Feb 24. J Cell Biol. 2014. PMID: 24567362 Free PMC article.
References
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. - PubMed
- Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10:675–678. - PubMed
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous