The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries - PubMed (original) (raw)
The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries
Miki Fujioka et al. Development. 2002 Oct.
Abstract
During segmentation of the Drosophila embryo, even skipped is required to activate engrailed stripes and to organize odd-numbered parasegments. A 16 kb transgene containing the even skipped coding region can rescue normal engrailed expression, as well as all other aspects of segmentation, in even skipped null mutants. To better understand its mechanism of action, we functionally dissected the Even-skipped protein in the context of this transgene. We found that Even-skipped utilizes two repressor domains to carry out its function. Each of these domains can function autonomously in embryos when fused with the Gal4 DNA-binding domain. A chimeric protein consisting only of the Engrailed repressor domain and the Even-skipped homeodomain, but not the homeodomain alone, was able to restore function, indicating that the repression of target genes is sufficient for even skipped function at the blastoderm stage, while the homeodomain is sufficient to recognize those target genes. When Drosophila Even skipped was replaced by its homologs from other species, including a mouse homolog, they could provide substantial function, indicating that these proteins can recognize similar target sites and also provide repressor activity. Using this rescue system, we show that broad, early even skipped stripes are sufficient for activation of both odd- and even-numbered engrailed stripes. Furthermore, these 'unrefined' stripes organize odd-numbered parasegments in a dose-dependent manner, while the refined, late stripes, which coincide cell-for-cell with parasegment boundaries, are required to ensure the stability of the boundaries.
Figures
Fig. 1
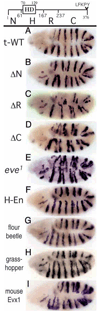
Functional dissection of Eve, and functional rescue by Eve homologues. At the top is a diagram of Drosophila Eve protein domains (for details see Material and Methods). The HD and Gro interaction domain (LFKPY) are indicated. Flag-tagged modified Eve proteins were expressed using the eve rescue construct (from −6.4 kb to +8.6 kb) in a Df(2R)eve mutant background. Patterns of expression of en mRNA were monitored by in situ hybridization. (A) Rescue by tagged, wild-type Eve; note the equally spaced en stripes. (B) The same construct with the N domain removed (ΔN); note that the spacing of en stripes is similar to wild type, although odd-numbered parasegments are slightly wider than even-numbered ones, indicating an increased activity. (C) The same construct with the R domain removed (ΔR); odd-numbered parasegments are severely narrowed and some odd-numbered stripes are missing, indicating a severe, but not complete, loss of activity. (D) LFKPY-deleted Eve (ΔC) (Kobayashi et al., 2001); the odd-numbered parasegments are narrowed, as in eve1 mutants (E) at a semi-permissive temperature (18°C), indicating a partial loss of activity. (F) A chimera of the Eve HD and En repressor domains (H-En); both en stripes and parasegment spacing are rescued. (G) Tc-Eve (flour beetle); all en stripes are restored, but odd-numbered parasegments are slightly narrowed. (H) Sa-Eve (grasshopper); all en stripes are restored, but odd-numbered parasegments are narrower. (I) Mouse Evx1; en stripes are restored, but spacing is abnormal, due to a combination of increased protein stability and variations in expression among the early stripes (Evx1 stripes 4, 5, and 6 are weak, and the corresponding parasegments, 7, 9 and 11, are narrowed, see text).
Fig. 2
Eve repressor domains function autonomously in embryos. (A–D) Expression patterns of a responder transgene (diagrammed at the top right) driven by the stripes 4+6, 1 and 5 enhancers and containing a Gal4-UAS sequence, visualized by in situ hybridization (to lacZ mRNA). (E–G) Expression patterns of the Gal4 fusion proteins indicated on the left, which contain Eve repressor regions, driven by the stripe 1 and 5 enhancers. The patterns in E and G were visualized by staining with polyclonal α-Eve antiserum, and that in F with monoclonal antibody 2B8, which specifically recognizes the Eve C terminus (see text), all in Df(2R)eve embryos. (A) The responder transgene alone, expressed in the pattern of eve stripes 1, 4, 5 and 6. Note that stripe 5 is slightly stronger than stripes 4 and 6. (B) The responder repressed by Gal4-Eve-RC, present in the stripe 1 and 5 regions, as shown in E (one copy of each transgene). Note that stripe 1 is dramatically reduced, while stripe 5 is slightly reduced relative to A. (C) The responder repressed by Gal4-Eve-C. Note that stripe 1 is strongly reduced, although not as much as in B, while stripe 5 is again slightly reduced. (D) The responder repressed by Gal4-Eve-R. Note that stripe 1 is clearly reduced, while stripe 5 may also be reduced.
Fig. 3
Early eve stripes set parasegment spacing and activate en, while the late stripes ensure maintenance of en expression and repression of slp. (A,B,D,J) Expression patterns of wg (blue) and En (orange), visualized by in situ hybridization and monoclonal (4D9) α-En staining. (C,G,H) Expression pattern of Eve, visualized by polyclonal α-Eve staining. (I) Expression pattern of en (blue) and Eve (orange), visualized by in situ hybridization and polyclonal α-Eve staining. (K,L) Expression pattern of slp, visualized by in situ hybridization. (A) An eve null mutant rescued by two copies of the wild-type rescue transgene (from −6.4 kb to +9.2 kb); both wg and En are expressed normally, and parasegments are equally spaced. (B) An eve null mutant rescued by a single copy of the same wild-type rescue transgene; note that the odd-numbered parasegments are severely narrowed (all wg and En stripes are expressed, but there are few if any non-_en/wg_-expressing cells in odd-numbered parasegments). (C) Early Eve stripe expression from a transgene that lacks the early stripe 4+6 enhancer (Δ46). (D) The embryonic phenotype of an eve null mutant rescued by Δ46; note that parasegments 7 and 11 (marked by bars) are severely narrowed, although en stripes 7 and 11 are expressed, at least at early stages (by this stage in this embryo, stripe 11 has almost faded). (E) Wild-type adult fly with normal segmentation. (F) The adult phenotype of an eve null mutant rescued by Δ46; note that there are two fewer abdominal segments. (G) Normal Eve expression, in Df(2R)eve rescued by two copies of the wild-type rescue transgene. On the right is a magnified view of the boxed region; note that the anterior (left) edge is sharply defined, with the anterior-most cell usually expressing the highest level. (H) Eve expression from two copies of a transgene that lacks the late element (Δlate), in a Df(2R)eve background; note that there is residual expression from early stripes, but that the high level expression at the anterior edge of each early stripe is missing. On the right is a magnified view of the boxed region; note that the anterior edge is less sharply defined than in G, that the stripe appears broader, and that the anterior-most cell row is not usually the highest expressing. The embryo in H is actually over-stained relative to that in G, as suggested by the fact that the stripes appear narrower in G, owing to a lack of detection of the low level expression in the posterior of each stripe. (I) The Δlate rescued phenotype early in gastrulation: odd-numbered en stripes are activated normally (marked by dots); note the regular parasegment spacing (except for parasegment 3, which is slightly narrower due to weaker than normal expression of early Eve stripe 3 in this line). (J) The Δlate rescued phenotype during germ band extension: odd-numbered en stripes are either narrowed or lost (marked by dots), and some wg stripes are expanded posteriorly. (K) Expression pattern of slp (indistinguishable from wild type) in an eve null mutant rescued by the wild-type rescue transgene. (L) Expression pattern of slp in a Δlate-rescued _eve_null; note that in even-numbered parasegments, slp is expanded posteriorly (into the regions of odd-numbered en stripes, marked by dots).
Fig. 4
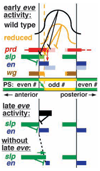
Model of Eve repressor function in segmentation. Early Eve activity establishes en expression in the proper positions. Reduction of Eve concentration or activity in the syncytial blastoderm (top half of figure; a gradient of both wild-type and reduced eve activity is diagrammed) reduces repression of at least two key target genes, slp and prd. Concentration-dependent effects at the anterior edge of each Eve stripe include expanded prd and slp expression (lighter colored bars) (see also Fujioka et al., 1995; Kobayashi et al., 2001). Prd activates both en and wg, while slp represses en. Thus slp (and possibly other en repressors that are repressed by Eve) can effectively subdivide the prd domain into _wg_- and _en_-expressing cells (en and eve repress wg). The border between slp and en becomes the parasegment boundary, and the overall width of the parasegment is largely determined by the location of this border. A dotted line indicates the shifted position of the parasegment boundary when early eve activity is reduced. The net effect of reducing early eve activity is to reduce the width of the odd-numbered parasegments, and to sometimes expand the odd-numbered en stripes, since prd sometimes expands more than does slp. There may also be effects at the posterior border of each early eve stripe, but these appear to be relatively minor. For example, ftz stripe 4 expression does not appreciably expand or shift in the absence of eve stripe 4 (Fujioka et al., 1995), although there may be an anterior shift of some ftz stripes in the absence of eve (see also Hughes and Krause, 2001). PS, parasegments; ⊥ indicates repression of target genes. Late Eve expression is required to maintain en expression. The absence of late eve activity (bottom half of figure) results in the expansion of slp expression, and the concomitant loss of en expression, beginning with the anterior of each en stripe. The continued presence of Eve just posterior to slp is thus necessary to prevent ‘encroachment’ of slp into the en stripe, and disruption of the incipient parasegment boundary.
Similar articles
- Drawing lines in the sand: even skipped et al. and parasegment boundaries.
Jaynes JB, Fujioka M. Jaynes JB, et al. Dev Biol. 2004 May 15;269(2):609-22. doi: 10.1016/j.ydbio.2004.03.001. Dev Biol. 2004. PMID: 15110723 Free PMC article. - Control of segmental asymmetry in Drosophila embryos.
Manoukian AS, Krause HM. Manoukian AS, et al. Development. 1993 Jul;118(3):785-96. doi: 10.1242/dev.118.3.785. Development. 1993. PMID: 7915670 - Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients.
Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Fujioka M, et al. Development. 1999 Jun;126(11):2527-38. doi: 10.1242/dev.126.11.2527. Development. 1999. PMID: 10226011 Free PMC article. - Establishing parasegments in Drosophila embryos: roles of the odd-skipped and naked genes.
Mullen JR, DiNardo S. Mullen JR, et al. Dev Biol. 1995 May;169(1):295-308. doi: 10.1006/dbio.1995.1145. Dev Biol. 1995. PMID: 7750646 - Cell patterning in the Drosophila segment: engrailed and wingless antigen distributions in segment polarity mutant embryos.
van den Heuvel M, Klingensmith J, Perrimon N, Nusse R. van den Heuvel M, et al. Dev Suppl. 1993:105-14. Dev Suppl. 1993. PMID: 8049466 Review.
Cited by
- Sex-specific pattern formation during early Drosophila development.
Manu, Ludwig MZ, Kreitman M. Manu, et al. Genetics. 2013 May;194(1):163-73. doi: 10.1534/genetics.112.148205. Epub 2013 Feb 14. Genetics. 2013. PMID: 23410834 Free PMC article. - Quantitative analysis reveals genotype- and domain- specific differences between mRNA and protein expression of segmentation genes in Drosophila.
Surkova S, Sokolkova A, Kozlov K, Nuzhdin SV, Samsonova M. Surkova S, et al. Dev Biol. 2019 Apr 1;448(1):48-58. doi: 10.1016/j.ydbio.2019.01.006. Epub 2019 Jan 7. Dev Biol. 2019. PMID: 30629954 Free PMC article. - Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness.
Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Ludwig MZ, et al. PLoS Genet. 2011 Nov;7(11):e1002364. doi: 10.1371/journal.pgen.1002364. Epub 2011 Nov 10. PLoS Genet. 2011. PMID: 22102826 Free PMC article. - Lack of tailless leads to an increase in expression variability in Drosophila embryos.
Janssens H, Crombach A, Wotton KR, Cicin-Sain D, Surkova S, Lim CL, Samsonova M, Akam M, Jaeger J. Janssens H, et al. Dev Biol. 2013 May 1;377(1):305-17. doi: 10.1016/j.ydbio.2013.01.010. Epub 2013 Jan 18. Dev Biol. 2013. PMID: 23333944 Free PMC article. - How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation.
Schroeder MD, Greer C, Gaul U. Schroeder MD, et al. Development. 2011 Jul;138(14):3067-78. doi: 10.1242/dev.062141. Development. 2011. PMID: 21693522 Free PMC article.
References
- Ahringer J. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 1996;10:1120–1130. - PubMed
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
- Brown SJ, Parrish JK, Beeman RW, Denell RE. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech. Dev. 1997;61:165–173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous