Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease - PubMed (original) (raw)
. 2002 Sep 15;22(18):7862-72.
doi: 10.1523/JNEUROSCI.22-18-07862.2002.
Lisa M Ellerby, Claire-Anne Gutekunst, Danny Rogers, Simon Warby, Rona K Graham, Odell Loubser, Jeremy van Raamsdonk, Roshni Singaraja, Yu-Zhou Yang, Juliette Gafni, Dale Bredesen, Steven M Hersch, Blair R Leavitt, Sophie Roy, Donald W Nicholson, Michael R Hayden
Affiliations
- PMID: 12223539
- PMCID: PMC6758089
- DOI: 10.1523/JNEUROSCI.22-18-07862.2002
Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease
Cheryl L Wellington et al. J Neurosci. 2002.
Abstract
Huntington's disease (HD) results from polyglutamine expansion in huntingtin (htt), a protein with several consensus caspase cleavage sites. Despite the identification of htt fragments in the brain, it has not been shown conclusively that htt is cleaved by caspases in vivo. Furthermore, no study has addressed when htt cleavage occurs with respect to the onset of neurodegeneration. Using antibodies that detect only caspase-cleaved htt, we demonstrate that htt is cleaved in vivo specifically at the caspase consensus site at amino acid 552. We detect caspase-cleaved htt in control human brain as well as in HD brains with early grade neuropathology, including one homozygote. Cleaved htt is also seen in wild-type and HD transgenic mouse brains before the onset of neurodegeneration. These results suggest that caspase cleavage of htt may be a normal physiological event. However, in HD, cleavage of mutant htt would release N-terminal fragments with the potential for increased toxicity and accumulation caused by the presence of the expanded polyglutamine tract. Furthermore, htt fragments were detected most abundantly in cortical projection neurons, suggesting that accumulation of expanded htt fragments in these neurons may lead to corticostriatal dysfunction as an early event in the pathogenesis of HD.
Figures
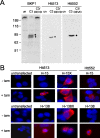
Fig. 1.
Validation of cleavage-specific antibodies Htt513 and Htt552. A, Western blot of wild-type murine cortical tissue demonstrating detection of a single htt fragment ending at either amino acid 513 or 552 that is present only after ex vivo cleavage of endogenous htt with recombinant caspase-3.B, Immunofluorescence of HEK 293T cells transfected with htt constructs as indicated, demonstrating that the Htt513 or Htt552 antibodies detect htt only after tamoxifen (tam)-induced cleavage or, for Htt513, in the presence of constructs terminating at amino acid 513 (H-15X), in red. Nuclei are counterstained with DAPI (blue).
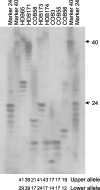
Fig. 2.
Determination of the CAG repeat length in HD brains (HDB) and control brains (COB). PCR products containing the htt CAG repeat were resolved on 0.5 mm sequencing gels alongside markers with a known CAG size. The total CAG size detected in the upper and lower alleles is listed below each_lane_.
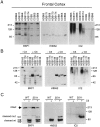
Fig. 3.
Htt is cleaved at amino acid 552 in vivo. A, Parallel blots prepared from human cortex homogenized in the presence of zVAD-fmk were probed with BKP1, Htt552, and 1C2. Cleavage products showing CAG-dependent mobility are detected by BKP1 (left panel) in all samples except HD brain (HDB) 174 (in this exposure) and by Htt552 (center panel) but not 1C2 (right panel). B, Human postmortem tissue was homogenized in the absence of zVAD-fmk and incubated at 37°C with or without 2 μ
m
recombinant caspase-3. Replicate blots were probed with BKP1 (left panel), Htt552 (center panel), or 1C2 (right panel). Caspase cleavage products arising from wild-type or expanded human htt in human brain lysates are detected with BKP1 and Htt552 but not 1C2. COB, Control brain.C, Whole murine brain from 6-month-old wild-type (WT) or YAC72 (line 2511) transgenic mice was homogenized in the absence of zVAD-fmk and incubated at 37°C with (+) or without (−) caspase-3 (C3). Replicate blots were probed with BKP1 (left panel), Htt552 (center panel), or 1C2 (right panel). Caspase cleavage products arising from wild-type murine (double open arrow) or expanded human htt (double asterisk) are distinguishable by the polyglutamine-mediated mobility shift. Full-length htt as well as normal and expanded fragments cleaved at amino acid 552 (upper band per allele) or amino acid 513 (lower band per allele) are detected with BKP1 (left panel). Htt552 recognizes only htt cleaved at amino acid 552 with normal and expanded polyglutamine (center panel). 1C2 detects both caspase cleavage products generated from the expanded human htt transgene in the context of murine brain. Faint immunoreactivity to intact expanded htt is also evident (left panel).
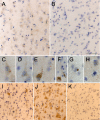
Fig. 4.
Immunohistochemical analysis of htt cleavage. A, Htt552 immunostaining of frontal cortex from control brain showing Htt552 immunoreactivity in many but not all pyramidal cortical neurons. B, Control panel for Htt552 immunostaining on the same control brain showing no immunoreactivity in the absence of primary antibody. Scale bar, 100 μm.C–H, High-power magnification of Htt552 immunostaining on frontal cortex from the grade I homozygote HDB171, showing the punctate, predominately cytoplasmic staining evident in cortical projection neurons. Scale bars: (in C) C–E,G, H, 20 μm; F, 10 μm.I, Htt552 immunostaining of frontal cortex from a 32-year-old presymptomatic patient (HDB Emory) showing the presence of cleaved htt in some neurons in layers II and III. J, Htt552 staining of frontal cortex from the same presymptomatic patient (HDB Emory) showing abundant immunoreactivity in pyramidal cortical neurons of layers V and VI. K, Htt552 immunostaining on striatum from the same presymptomatic patient, showing that some striatal neurons contain caspase-cleaved htt. Scale bar, 100 μm.
Fig. 5.
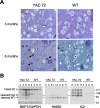
Degeneration and cleavage of htt in YAC transgenic mice with 72 CAG repeats. A, Semithin striatal sections from wild-type (WT) and YAC72 (line 44) transgenic mice stained with toluidine blue. Healthy neurons appear clear (white arrowhead), and degenerating neurons appear darkly stained (black arrows). B, Western blots of cortex from 2-, 4-, 6-, and 11-month-old YAC72 (line 44) transgenic mice or wild-type littermate controls. Blots were probed with BKP1 (top portion) and GAPDH (bottom portion) for a loading control (left panel), Htt552 (center panel), and 1C2 (right panel). Intact htt is indicated by an_open arrowhead_ and is detected by BKP1 (normal and expanded) and by 1C2 (expanded). The specific 100 and 70 kDa caspase cleavage products of expanded (cleaved mut) and normal (cleaved WT) alleles are indicated by the_closed_ and open arrows, respectively. Expanded cleaved htt is detected by all three antibodies, where as only BKP1 and Htt552 detect cleaved htt with a normal CAG repeat.
Fig. 6.
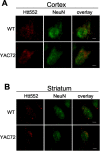
Immunofluorescence of 2-month-old YAC72 (line 44) and wild-type (WT) mice. A, In the cortex, Htt552 immunostaining (red) appears in a punctate pattern throughout the neuron, as confirmed by costaining with the neuronal marker NeuN (green).B, Immunofluorescence of the striatum from YAC72 (line 44) and wild-type mice at 2 months of age. Htt552 immunostaining (red) appears in a punctate pattern throughout the neuron, as confirmed by costaining with the neuronal marker NeuN (green). Htt552 staining was more abundant in the cortex than in the striatum. Scale bars, 2.5 μm.
Fig. 7.
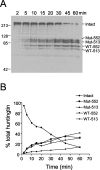
Kinetic analysis of htt cleavage in brain homogenates. A, YAC72 (high expressing) whole-brain homogenate prepared in the absence of zVAD-fmk was incubated for various times with a final concentration of 2 n
m
caspase-3, followed by immunodetection of cleavage fragments with BKP1. Over time, total intact htt (Intact) is converted to four major cleavage products, including mutant transgenic htt cleaved at amino acid 552 (Mut-552), mutant transgenic htt cleaved at amino acid 513 (Mut-513), wild-type endogenous htt cleaved at amino acid 552 (WT-552), and wild-type endogenous htt cleaved at amino acid 513 (WT-513).B, Quantitation of the percentage of reduction of intact total htt and percentage of accumulation of each of the htt cleavage products shows that mutant and wild-type cleavage products accumulate at equivalent rates.
Similar articles
- Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington's disease.
Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, Hackam AS, Logvinova AV, Peel AL, Chen SF, Hook V, Singaraja R, Krajewski S, Goldsmith PC, Ellerby HM, Hayden MR, Bredesen DE, Ellerby LM. Hermel E, et al. Cell Death Differ. 2004 Apr;11(4):424-38. doi: 10.1038/sj.cdd.4401358. Cell Death Differ. 2004. PMID: 14713958 - Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin.
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR. Graham RK, et al. Cell. 2006 Jun 16;125(6):1179-91. doi: 10.1016/j.cell.2006.04.026. Cell. 2006. PMID: 16777606 - Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis.
Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. Kim YJ, et al. Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12784-9. doi: 10.1073/pnas.221451398. Proc Natl Acad Sci U S A. 2001. PMID: 11675509 Free PMC article. - Huntington disease: new insights on the role of huntingtin cleavage.
Wellington CL, Leavitt BR, Hayden MR. Wellington CL, et al. J Neural Transm Suppl. 2000;(58):1-17. doi: 10.1007/978-3-7091-6284-2_1. J Neural Transm Suppl. 2000. PMID: 11128600 Review. - Selective degeneration in YAC mouse models of Huntington disease.
Van Raamsdonk JM, Warby SC, Hayden MR. Van Raamsdonk JM, et al. Brain Res Bull. 2007 Apr 30;72(2-3):124-31. doi: 10.1016/j.brainresbull.2006.10.018. Epub 2006 Nov 16. Brain Res Bull. 2007. PMID: 17352936 Review.
Cited by
- Protective Proteolysis in Huntington's Disease: Unraveling the Role of Post-Translational Myristoylation of Huntingtin in Autophagy.
Alshehabi Y, Martin DDO. Alshehabi Y, et al. J Huntingtons Dis. 2024;13(3):267-277. doi: 10.3233/JHD-240028. J Huntingtons Dis. 2024. PMID: 38995796 Free PMC article. Review. - Downregulation of glial genes involved in synaptic function mitigates Huntington's disease pathogenesis.
Onur TS, Laitman A, Zhao H, Keyho R, Kim H, Wang J, Mair M, Wang H, Li L, Perez A, de Haro M, Wan YW, Allen G, Lu B, Al-Ramahi I, Liu Z, Botas J. Onur TS, et al. Elife. 2021 Apr 19;10:e64564. doi: 10.7554/eLife.64564. Elife. 2021. PMID: 33871358 Free PMC article. - The Novel Alpha-2 Adrenoceptor Inhibitor Beditin Reduces Cytotoxicity and Huntingtin Aggregates in Cell Models of Huntington's Disease.
Singer E, Hunanyan L, Melkonyan MM, Weber JJ, Danielyan L, Nguyen HP. Singer E, et al. Pharmaceuticals (Basel). 2021 Mar 12;14(3):257. doi: 10.3390/ph14030257. Pharmaceuticals (Basel). 2021. PMID: 33809220 Free PMC article. - The Emerging Landscape of Natural Small-molecule Therapeutics for Huntington's Disease.
Bhat SA, Ahamad S, Dar NJ, Siddique YH, Nazir A. Bhat SA, et al. Curr Neuropharmacol. 2023;21(4):867-889. doi: 10.2174/1570159X21666230216104621. Curr Neuropharmacol. 2023. PMID: 36797612 Free PMC article. Review. - A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila.
Apostol BL, Kazantsev A, Raffioni S, Illes K, Pallos J, Bodai L, Slepko N, Bear JE, Gertler FB, Hersch S, Housman DE, Marsh JL, Thompson LM. Apostol BL, et al. Proc Natl Acad Sci U S A. 2003 May 13;100(10):5950-5. doi: 10.1073/pnas.2628045100. Epub 2003 May 1. Proc Natl Acad Sci U S A. 2003. PMID: 12730384 Free PMC article.
References
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. - PubMed
- Bruland O, Almqvist EW, Goldberg YP, Boman H, Hayden MR, Knappskog PM. Accurate determination of the number of CAG repeats in the Huntington disease gene using a sequence-specific internal DNA standard. Clin Genet. 1999;55:198–202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS40251/NS/NINDS NIH HHS/United States
- R01 AT000613/AT/NCCIH NIH HHS/United States
- NS35255/NS/NINDS NIH HHS/United States
- R01 NS040251/NS/NINDS NIH HHS/United States
- U01 AT000613/AT/NCCIH NIH HHS/United States
- AT00613/AT/NCCIH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous