The circadian clock that controls gene expression in Arabidopsis is tissue specific - PubMed (original) (raw)
Comparative Study
. 2002 Sep;130(1):102-10.
doi: 10.1104/pp.005405.
Affiliations
- PMID: 12226490
- PMCID: PMC166543
- DOI: 10.1104/pp.005405
Comparative Study
The circadian clock that controls gene expression in Arabidopsis is tissue specific
Simon C Thain et al. Plant Physiol. 2002 Sep.
Abstract
The expression of CHALCONE SYNTHASE (CHS) expression is an important control step in the biosynthesis of flavonoids, which are major photoprotectants in plants. CHS transcription is regulated by endogenous programs and in response to environmental signals. Luciferase reporter gene fusions showed that the CHS promoter is controlled by the circadian clock both in roots and in aerial organs of transgenic Arabidopsis plants. The period of rhythmic CHS expression differs from the previously described rhythm of chlorophyll a/b-binding protein (CAB) gene expression, indicating that CHS is controlled by a distinct circadian clock. The difference in period is maintained in the wild-type Arabidopsis accessions tested and in the de-etiolated 1 and timing of CAB expression 1 mutants. These clock-affecting mutations alter the rhythms of both CAB and CHS markers, indicating that a similar (if not identical) circadian clock mechanism controls these rhythms. The distinct tissue distribution of CAB and CHS expression suggests that the properties of the circadian clock differ among plant tissues. Several animal organs also exhibit heterogeneous circadian properties in culture but are believed to be synchronized in vivo. The fact that differing periods are manifest in intact plants supports our proposal that spatially separated copies of the plant circadian clock are at most weakly coupled, if not functionally independent. This autonomy has apparently permitted tissue-specific specialization of circadian timing.
Figures
Figure 1
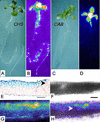
Tissue-specific expression of the CHS promoter. Reflected-light (A and C) and luminescence (B and D) images of CHS:LUC (A and B) and CAB:LUC (C and D) seedlings after 12 d of growth in LD (12, 12) of 150 μmol m−2 s−1. Luminescence video images were processed in false color (see “Materials and Methods”): White and red shades represent the highest photon counts, and darker blues, the lowest. A reflected-light image of the plant is shown to the left of each false color image. E through H, Leaf sections were prepared from plants grown in LD (12, 12) of 150 μmol m−2 s−1 for 7 d and then transferred to 250 μmol m−2 s−1 for 5 d. E, Microtome section of a CHS:β-glucuronidase (GUS) leaf, stained for GUS activity and counter-stained with ruthenium red. F through H, Thick hand section of a CHS:LUC leaf: bright field (F), luminescence (G), and overlaid (H) images. E and F, Scale bars represent 60 μm. E and H, Arrowheads indicate areas where CHS expression is very weak or absent from the mesophyll.
Figure 2
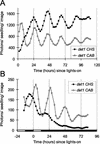
Rhythmic luminescence in roots and aerial organs of Arabidopsis seedlings expressing CHS:LUC. CHS:Luc (A, B, C, and E) or mCHS:LUC (D) expression was assayed by video imaging (see “Materials and Methods”) at the times indicated. A, B, D, and E, Seedlings were grown on vertical agar plates, in 7 d of LD (12, 12) at 150 μmol m−2 s−1. Seedlings in A and D were transferred (at 0 h) to continuous light of 150 μmol m−2 s−1; seedlings in B were transferred to continuous light of 250 μmol m−2 s−1; and seedlings in E were transferred first to 250 μmol m−2 s−1 for 12 h and then to constant darkness. Seedlings in C were both grown and tested at 250 μmol m−2 s−1. The data are representative of at least five independent experiments. White box on time axis, Light interval; black box, dark interval. Lv, Luminescence from aerial organs; Rt, luminescence from roots.
Figure 3
Circadian rhythms of CHS:LUC activity in det1. Expression was assayed as in Figure 2. Seedlings were grown in LD (8, 16) at 20 μmol m−2 s−1 to confirm the det1 phenotype. Seedlings were transferred to LD (12, 12) at 150 μmol m−2 s−1, 2 d before the start of the experiment and to constant light of 250 μmol m−2 s−1 at 0 h (A) or to 250 μmol m−2 s−1 at 0 h and constant darkness at 12 h (B). Luminescence data were analyzed from whole seedlings, without separate analysis of roots and aerial tissues. The data are representative of at least four independent experiments. White box on time axis, Light interval; black box, dark interval.
Figure 4
Autonomy and specialization of the circadian clock. A simplified model of the circadian clock is shown in the mesophyll and in the epidermis, with the output markers and clock-related genes tested in this work. The clock is depicted as containing the same components in each case; TOC1 is thought to function in the oscillator, and DET1 is thought to affect the light input pathway from the photoreceptors. Rhythmic output from the oscillator is shown regulating transcription factors at the CAB or CHS promoter. Two hypothetical tissue-specific factors are shown: modification of the light input pathway and an unknown tissue-specific factor (X). One or both modify the function of an oscillator component, resulting in a longer period in the epidermis.
Similar articles
- Conditional circadian regulation of PHYTOCHROME A gene expression.
Hall A, Kozma-Bognár L, Tóth R, Nagy F, Millar AJ. Hall A, et al. Plant Physiol. 2001 Dec;127(4):1808-18. Plant Physiol. 2001. PMID: 11743124 Free PMC article. - Circadian clock mutants in Arabidopsis identified by luciferase imaging.
Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Millar AJ, et al. Science. 1995 Feb 24;267(5201):1161-3. doi: 10.1126/science.7855595. Science. 1995. PMID: 7855595 - DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock.
Song HR, Carré IA. Song HR, et al. Plant Mol Biol. 2005 Mar;57(5):761-71. doi: 10.1007/s11103-005-3096-z. Plant Mol Biol. 2005. PMID: 15988568 - Shedding light on clock controlled cab gene transcription in higher plants.
Kay SA. Kay SA. Semin Cell Biol. 1993 Apr;4(2):81-6. doi: 10.1006/scel.1993.1010. Semin Cell Biol. 1993. PMID: 8318699 Review. - How plants tell the time.
Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AA. Gardner MJ, et al. Biochem J. 2006 Jul 1;397(1):15-24. doi: 10.1042/BJ20060484. Biochem J. 2006. PMID: 16761955 Free PMC article. Review.
Cited by
- Unlocking Nature's Rhythms: Insights into Secondary Metabolite Modulation by the Circadian Clock.
Pérez-Llorca M, Müller M. Pérez-Llorca M, et al. Int J Mol Sci. 2024 Jul 3;25(13):7308. doi: 10.3390/ijms25137308. Int J Mol Sci. 2024. PMID: 39000414 Free PMC article. Review. - Integrated Metabolomic-Transcriptomic Analyses of Flavonoid Accumulation in Citrus Fruit under Exogenous Melatonin Treatment.
Zhao C, Wang Z, Liao Z, Liu X, Li Y, Zhou C, Sun C, Wang Y, Cao J, Sun C. Zhao C, et al. Int J Mol Sci. 2024 Jun 16;25(12):6632. doi: 10.3390/ijms25126632. Int J Mol Sci. 2024. PMID: 38928338 Free PMC article. - Overexpression of a pseudo-etiolated-in-light-like protein in Taraxacum koksaghyz leads to a pale green phenotype and enables transcriptome-based network analysis of photomorphogenesis and isoprenoid biosynthesis.
Wolters SM, Benninghaus VA, Roelfs KU, van Deenen N, Twyman RM, Prüfer D, Schulze Gronover C. Wolters SM, et al. Front Plant Sci. 2023 Sep 28;14:1228961. doi: 10.3389/fpls.2023.1228961. eCollection 2023. Front Plant Sci. 2023. PMID: 37841614 Free PMC article. - A wound inducible chalcone synthase gene from Dysoxylum gotadhora (DbCHS) regulates flavonoid biosynthesis.
Mahajan V, Chouhan R, Jamwal VL, Kapoor N, Gandhi SG. Mahajan V, et al. Physiol Mol Biol Plants. 2023 Jul;29(7):959-969. doi: 10.1007/s12298-023-01344-2. Epub 2023 Aug 21. Physiol Mol Biol Plants. 2023. PMID: 37649885 Free PMC article. - Limited water stress modulates expression of circadian clock genes in Brachypodium distachyon roots.
Gombos M, Hapek N, Kozma-Bognár L, Grezal G, Zombori Z, Kiss E, Györgyey J. Gombos M, et al. Sci Rep. 2023 Jan 23;13(1):1241. doi: 10.1038/s41598-022-27287-4. Sci Rep. 2023. PMID: 36690685 Free PMC article.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1. Science. 2001;293:880–883. - PubMed
- Batschauer A, Ehmann B, Schafer E. Cloning and characterization of a chalcone synthase gene from mustard and its light-dependent expression. Plant Mol Biol. 1991;16:175–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases