Nodal activity in the node governs left-right asymmetry - PubMed (original) (raw)
Nodal activity in the node governs left-right asymmetry
Jane Brennan et al. Genes Dev. 2002.
Abstract
Nodal is expressed at the lateral edges of the mouse node, but its function in this "organizer" tissue remains unknown due to the early lethality of Nodal mutant embryos. Here we used a genetic strategy to selectively remove Nodal activity from the node. Embryos lacking Nodal in the node fail to initiate molecular asymmetry in the left lateral plate mesoderm and exhibit multiple left-right patterning defects. Nodal may also act as a short-range signal to establish a functional midline barrier. Our findings confirm that the mouse node is instrumental in initiating left-right axis specification and identify Nodal as the key morphogen regulating this process.
Figures
Figure 1
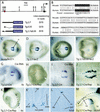
Characterization of the Nodal node enhancer. Ventral (C_–_J) and lateral (K) views of approximately 8.0-d embryos at the various somite (s) stages indicated. (A) Map of Nodal locus illustrating location of node enhancer element (red circle) and three LacZ reporter constructs used to make stable transgenic lines. Black box indicates position of exon 1 (K, Kpn I; B, Bgl II; N, Not I). (C) Nodal is expressed in the outermost ventral cells of the node at the 2-somite stage as assessed by the NodallacZ reporter allele. Transgenic lines containing 2.7 kb (A,D) and 0.35 kb (A,E) of Nodal genomic sequence express β-gal reporter in the outermost ventral cells of the node. Removal of the 0.35 kb from the 2.7-kb transgene eliminates β-gal expression from the outermost cells of the node (A,F); some staining is seen in the central cells of the node. (B) Partial sequence of the minimal 0.35-kb fragment that drives Nodal expression in the node. Alignment of mouse and human sequences showing predicted Foxa2 binding site. Mismatches between the second and third bases and the Foxa2 consensus are compatible with Foxa2 binding (Overdier et al. 1994). (G) Whole-mount in situ hybridization showing Cre expression in the node of a transgenic line expressing Cre under the control of Tg 2.7 (the 2.7-kb node element). (H) Embryo carrying the Tg 2.7-Cre transgene and an allele of the Rosa-26 conditional reporter. Only a few cells show β-gal activity in the node (white arrow). At the 3-somite stage, Cre mRNA is expressed in the node (I), and Cre activity is detected throughout the outermost ventral cells of the node as visualized by β-gal activity (J). By the 6-somite stage, descendants of _Nodal_-expressing cells in the node are found exclusively in the notochordal plate (K‘). By 9.5 d (L) and 10.5 d (M), the node descendants are found in the notochord posterior to the hindlimb. No descendants are found in the gut or floor plate.
Figure 2
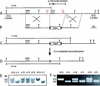
Targeted deletion of the Nodal node enhancer. (A) Map of Nodal genomic locus. (B) Targeting vector designed to replace 2.7 kb of Nodal sequence with a single loxP site (black triangle). (C) Targeted allele. (D) Recombined allele after Cre-mediated excision of the hygro selection cassette. (E) Southern hybridization of _Kpn_I-digested DNA prepared from tail biopsies of heterozygous intercross offspring at weaning. DNA was hybridized with a 5′ external probe (A). (F) PCR analysis of yolk sac DNA prepared from 8.0-dpc embryos. Primers C2/D2 amplify a wild-type band of 220 bp, and primers A3/D2 amplify a 320-bp mutant band. K, KpnI; B, BglII.
Figure 3
Left-right patterning defects of the viscera of _Nodal_Δ/− deletion mutants. Organs were dissected from control and deletion pups on the day of birth. Left (L) and right (R) are as indicated in A and B. Normally the stomach is situated on the left-hand side of the body (A), but in the deletion mutants the stomach position is randomized such that half the mutants have stomachs on the right (B). The spleen is apposed to the stomach wall (C). In mutants, the spleen is greatly reduced in size (D). In the normal lung, there is one lobe on the left and four on the right (E). In the mutants, there were three or four lobes on both sides, indicating a right pulmonary isomerism (F). At 9.5 d, expression of α-cardiac actin mRNA in the heart tube illustrates normal heart looping to the right (G). In _Nodal_Δ/− mutants, looping was randomized, with 3/7 showing rightward and 4/7 showing leftward looping (H). In normal littermates, the apex of the heart points to the left (I), yet in mutants the heart apex is ambiguously positioned such that a proportion point to the left, some to the middle (J), and others to the left. Sections through mutant hearts reveal septation defects between the two atria (J). A, accessory lobe; CA, common atrium; Ca, caudal lobe; Cr, cranial lobe; L, left lobe; LA, left atrium; LV, left ventricle; M, medial lobe; OT, outflow tract; RA, right atrium; RV, right ventricle; Vn, ventricles.
Figure 4
Molecular asymmetry is not established in the lateral plate mesoderm of _Nodal_Δ/− deletion mutants. Whole-mount in situ hybridization (A_–_C,F_–_L) and LacZ reporter staining (D,E) of 7.5-d (A_–_C) and 8.0-d (D_–_L) embryos. Lateral views with anterior to the left (A_–_C), caudal views (D_–_H), and rostral views (I_–_L) are shown. At the late head fold stage, Nodal is expressed in the node at the distal tip of the embryo (A); transverse sections through the node show that Nodal is expressed in the outermost ventral cells (A′;). In embryos homozygous for the NodalΔ/Δ allele, Nodal expression is downregulated in the node (B). In embryos trans-heterozygous for the deletion allele and a Nodal null allele (_Nodal_Δ/−), Nodal expression is either entirely absent or negligible in the node (C). Control embryos between 2 and 8 somites express the NodallacZ reporter in the node and the left LPM (D). In embryos carrying a copy of the deletion allele and the Nodal null reporter allele (_Nodal_Δ/LacZ), NodallacZ is only expressed in the node (E). Similarly Nodal mRNA is expressed in the node and left LPM in control embryos (F), but in NodalΔ/Δ homozygotes, low levels of Nodal are detected in the node yet normal levels of Nodal expression are observed in the left LPM (G). Reduced Nodal expression levels are also found in _Nodal_Δ/− nodes, and Nodal expression is absent from the left LPM (H‘). Lefty2 is expressed in the left LPM and the left prospective floor plate of control embryos (I) but fails to be induced in either domain in the deletion mutants (J). Pitx2 is expressed in the head mesenchyme, body wall, and left LPM of control embryos (K,K′;). In the deletion mutants, Pitx2 is expressed in the head mesenchyme and body wall (L) but is absent from the left LPM (L‘). Normal expression of Pitx2 in the left-hand side mesenchyme of the developing heart (arrow in K") is absent in the deletion mutants (L"). llpm, left lateral plate mesoderm; n, node; nf, neural fold; nt, neural tube; pfp, prospective floor plate. Black arrows indicate Nodal expression domain.
Similar articles
- Reversal of left-right asymmetry induced by aberrant Nodal signaling in the node of mouse embryos.
Oki S, Kitajima K, Marques S, Belo JA, Yokoyama T, Hamada H, Meno C. Oki S, et al. Development. 2009 Dec;136(23):3917-25. doi: 10.1242/dev.039305. Development. 2009. PMID: 19906859 - Cell fate decisions within the mouse organizer are governed by graded Nodal signals.
Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Vincent SD, et al. Genes Dev. 2003 Jul 1;17(13):1646-62. doi: 10.1101/gad.1100503. Genes Dev. 2003. PMID: 12842913 Free PMC article. - BMP antagonism is required in both the node and lateral plate mesoderm for mammalian left-right axis establishment.
Mine N, Anderson RM, Klingensmith J. Mine N, et al. Development. 2008 Aug;135(14):2425-34. doi: 10.1242/dev.018986. Epub 2008 Jun 11. Development. 2008. PMID: 18550712 - Establishment of vertebrate left-right asymmetry.
Hamada H, Meno C, Watanabe D, Saijoh Y. Hamada H, et al. Nat Rev Genet. 2002 Feb;3(2):103-13. doi: 10.1038/nrg732. Nat Rev Genet. 2002. PMID: 11836504 Review. - A conserved role for the nodal signaling pathway in the establishment of dorso-ventral and left-right axes in deuterostomes.
Duboc V, Lepage T. Duboc V, et al. J Exp Zool B Mol Dev Evol. 2008 Jan 15;310(1):41-53. doi: 10.1002/jez.b.21121. J Exp Zool B Mol Dev Evol. 2008. PMID: 16838294 Review.
Cited by
- Modeling Mammalian Commitment to the Neural Lineage Using Embryos and Embryonic Stem Cells.
Shparberg RA, Glover HJ, Morris MB. Shparberg RA, et al. Front Physiol. 2019 Jul 11;10:705. doi: 10.3389/fphys.2019.00705. eCollection 2019. Front Physiol. 2019. PMID: 31354503 Free PMC article. Review. - Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry.
Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Takeuchi JK, et al. Proc Natl Acad Sci U S A. 2007 Jan 16;104(3):846-51. doi: 10.1073/pnas.0608118104. Epub 2007 Jan 8. Proc Natl Acad Sci U S A. 2007. PMID: 17210915 Free PMC article. - A null allele of Dnaaf2 displays embryonic lethality and mimics human ciliary dyskinesia.
Cheong A, Degani R, Tremblay KD, Mager J. Cheong A, et al. Hum Mol Genet. 2019 Aug 15;28(16):2775-2784. doi: 10.1093/hmg/ddz106. Hum Mol Genet. 2019. PMID: 31107948 Free PMC article. - Pax9 and Gbx2 Interact in the Pharyngeal Endoderm to Control Cardiovascular Development.
Stothard CA, Mazzotta S, Vyas A, Schneider JE, Mohun TJ, Henderson DJ, Phillips HM, Bamforth SD. Stothard CA, et al. J Cardiovasc Dev Dis. 2020 May 25;7(2):20. doi: 10.3390/jcdd7020020. J Cardiovasc Dev Dis. 2020. PMID: 32466118 Free PMC article. - Nature and extent of left/right axis defects in T(Wis) /T(Wis) mutant mouse embryos.
Concepcion D, Papaioannou VE. Concepcion D, et al. Dev Dyn. 2014 Aug;243(8):1046-53. doi: 10.1002/dvdy.24144. Epub 2014 May 26. Dev Dyn. 2014. PMID: 24801048 Free PMC article.
References
- Beddington RSP. Induction of a second neural axis by the mouse node. Development. 1994;120:613–620. - PubMed
- Beddington RSP, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. - PubMed
- Bellomo D, Lander A, Harragan I, Brown NA. Cell proliferation in mammalian gastrulation: The ventral node and notochord are relatively quiescent. Dev Dyn. 1996;205:471–485. - PubMed
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RSP, Robertson EJ. Nodal signaling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. - PubMed
- Capdevila J, Vogan KJ, Tabin CJ, Izpisua Belmonte JC. Mechanisms of left-right determination in vertebrates. Cell. 2000;101:9–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases