Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor - PubMed (original) (raw)
Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor
Sanjay K Mishra et al. EMBO J. 2002.
Abstract
Clathrin-coated pits at the cell surface select material for transportation into the cell interior. A major mode of cargo selection at the bud site is via the micro 2 subunit of the AP-2 adaptor complex, which recognizes tyrosine-based internalization signals. Other internalization motifs and signals, including phosphorylation and ubiquitylation, also tag certain proteins for incorporation into a coated vesicle, but the mechanism of selection is unclear. Disabled-2 (Dab2) recognizes the FXNPXY internalization motif in LDL-receptor family members via an N-terminal phosphotyrosine-binding (PTB) domain. Here, we show that in addition to binding AP-2, Dab2 also binds directly to phosphoinositides and to clathrin, assembling triskelia into regular polyhedral coats. The FXNPXY motif and phosphoinositides contact different regions of the PTB domain, but can stably anchor Dab2 to the membrane surface, while the distal AP-2 and clathrin-binding determinants regulate clathrin lattice assembly. We propose that Dab2 is a typical member of a growing family of cargo-specific adaptor proteins, including beta-arrestin, AP180, epsin, HIP1 and numb, which regulate clathrin-coat assembly at the plasma membrane by synchronizing cargo selection and lattice polymerization events.
Figures
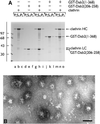
Fig. 1. Dab2 binds to both the AP-2 adaptor complex and clathrin. (A) Schematic illustration of the overall domain organization of Dab2 and the various GST–Dab2 constructs used. Phosphoinositide-, AP-2- and clathrin-binding properties of each fusion protein are indicated qualitatively. (B) Approximately 50 µg of either GST (lanes a and b), GST–Dab2(206–350) (lanes c and d), GST–Dab2(206–368) (lanes e and f) or GST–Dab2(206–492) (lanes g and h) immobilized on GSH–Sepharose were incubated with rat brain cytosol. After centrifugation, aliquots corresponding to 1/60 of each supernatant (S) and 1/5 of each washed pellet (P) were resolved by SDS–PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with the anti-AP-2 α-subunit mAb 100/2, anti-AP-2 µ2-subunit antiserum, the anti-clathrin heavy chain (HC) mAb TD.1 or the anti-clathrin light chain (LC) mAb Cl 57.3. The position of the molecular mass standards (in kDa) is indicated on the left and only the relevant portion of each blot is shown.
Fig. 2. A clathrin-binding region within Dab2. Approximately 50 µg of either GST (lanes a and b) or GST–Dab2(206–258) (lanes c and d), GST–Dab2(206–258) (LVD→AAA) (lanes e and f), GST–Dab2(206–368) (lanes g and h), GST–Dab2(206–368) (LVD→AAA) (lanes i and j) or GST–Dab2(206–368) (LVD→AAA/W→A) (lanes k and l) immobilized on GSH–Sepharose were incubated with rat brain cytosol. After centrifugation, aliquots corresponding to 1/50 of each supernatant (S) and 1/5 of each washed pellet (P) were resolved by SDS–PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with the anti-AP-2 α-subunit mAb 100/2, anti-AP-2 µ2-subunit antiserum, the anti-clathrin HC mAb TD.1 or the anti-clathrin LC mAb Cl 57.3.
Fig. 3. Dab2 associates directly with assembled clathrin cages. Pre-assembled clathrin cages (∼0.5 µM), GST, GST–Dab2(206–258), GST–Dab2(206–350) (each ∼1 µM) or combinations thereof were incubated in MES–OH buffer on ice. After centrifugation, aliquots corresponding to 1/10 of each supernatant (S) or 1/8 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue.
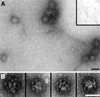
Fig. 4. Dab2 assembles soluble clathrin trimers into spherical polyhedral cages. (A) Clathrin (∼0.2 µM), GST–Dab2(206–258) (∼1 µM), GST–Dab2(1–368) (∼1 µM) or combinations thereof were mixed in 0.5 M Tris–HCl pH 7.0 and then dialyzed overnight against assay buffer. The samples were subject to differential centrifugation generating a low-speed pellet (PL) containing aggregated material, a high-speed supernatant (SH) containing soluble protein, and a high-speed pellet (PH) with the sedimentable assemblies. (B) Representative EM micrograph of a negatively stained aliquot of the high-speed pellet obtained from an incubation of clathrin with GST–Dab2(1–368) after resuspension in assay buffer on ice. Bar = 100 nm.
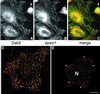
Fig. 5. Dab2 co-localizes with epsin 1 at the plasma membrane. Saponin-permeabilized HeLa cells were fixed and probed with antibodies against Dab2 (A) and epsin 1 (B). The merged color images (C–E) show that areas of overlap are extensive (yellow), but there are clear regions containing each protein alone. Single 0.3 µm confocal sections of the ventral surface (D) and the medial region of the cell (E) through the nucleus (N) show Dab2 co-localized with epsin at the cell surface (arrows). The arrowhead indicates a pre/post-mitotic cell where the Dab2 staining pattern is principally nuclear, although some epsin is still visible at the cell surface; we note that the intracellular distribution of Dab2 appears to alter with the cell cycle. Importantly, the anti-Dab2 mAb used here detects all three splice isoforms of Dab2 (p67, p93 and p96), so we cannot conclude whether the different locations represent distinct Dab2 isoforms. Bar = 25 µm in (A–C) and 10 µm in (D) and (E).
Fig. 6. The PTB domain of Dab2 binds to polyphosphoinositides. (A) GST–Dab2(1–205) (lanes a–f), GST–epsin 1(1–163) (lanes g and h) or GST (lanes i and j), as indicated, were mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated together on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. (B) Thrombin-cleaved Dab2(1–205) (lanes a–f) or GST (lanes g and h) was mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated on ice. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue or transferred to nitrocellulose. The blot was probed with an anti-Dab2 mAb. The asterisk denotes a stable ∼25 kDa thrombin degradation product that binds phosphoinositides and reacts with the anti-Dab2 mAb 52. (C) Nitrocellulose-immobilized phosphoinositides (100, 50, 25, 12.5, 6.2, 3.1, 1.5 pmol/spot) were incubated with GST–Dab2(1–205) PTB domain and then bound protein visualized with GSH-derivatized HRP. (D) The PTB domain of Dab2 binds to phosphoinositides and FXNPXF internalization motifs synchronously. Thrombin-cleaved Dab2(1–205) (PTB domain) was added to 3 µM, together with 0.4 mg/ml phosphoinositide-containing liposomes alone (lanes a and b) or liposomes plus either 15 µM NWRLKNINSIFDNPVYQKTT (lanes c and d) or NWRLKNINSIFDAPVAQKTT (lanes e and f) peptide and incubated on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. The bound peptide migrates at the dye front and the asterisk denotes the stable ∼25 kDa Dab2 degradation product of the PTB domain.
Fig. 6. The PTB domain of Dab2 binds to polyphosphoinositides. (A) GST–Dab2(1–205) (lanes a–f), GST–epsin 1(1–163) (lanes g and h) or GST (lanes i and j), as indicated, were mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated together on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. (B) Thrombin-cleaved Dab2(1–205) (lanes a–f) or GST (lanes g and h) was mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated on ice. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue or transferred to nitrocellulose. The blot was probed with an anti-Dab2 mAb. The asterisk denotes a stable ∼25 kDa thrombin degradation product that binds phosphoinositides and reacts with the anti-Dab2 mAb 52. (C) Nitrocellulose-immobilized phosphoinositides (100, 50, 25, 12.5, 6.2, 3.1, 1.5 pmol/spot) were incubated with GST–Dab2(1–205) PTB domain and then bound protein visualized with GSH-derivatized HRP. (D) The PTB domain of Dab2 binds to phosphoinositides and FXNPXF internalization motifs synchronously. Thrombin-cleaved Dab2(1–205) (PTB domain) was added to 3 µM, together with 0.4 mg/ml phosphoinositide-containing liposomes alone (lanes a and b) or liposomes plus either 15 µM NWRLKNINSIFDNPVYQKTT (lanes c and d) or NWRLKNINSIFDAPVAQKTT (lanes e and f) peptide and incubated on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. The bound peptide migrates at the dye front and the asterisk denotes the stable ∼25 kDa Dab2 degradation product of the PTB domain.
Fig. 6. The PTB domain of Dab2 binds to polyphosphoinositides. (A) GST–Dab2(1–205) (lanes a–f), GST–epsin 1(1–163) (lanes g and h) or GST (lanes i and j), as indicated, were mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated together on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. (B) Thrombin-cleaved Dab2(1–205) (lanes a–f) or GST (lanes g and h) was mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated on ice. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue or transferred to nitrocellulose. The blot was probed with an anti-Dab2 mAb. The asterisk denotes a stable ∼25 kDa thrombin degradation product that binds phosphoinositides and reacts with the anti-Dab2 mAb 52. (C) Nitrocellulose-immobilized phosphoinositides (100, 50, 25, 12.5, 6.2, 3.1, 1.5 pmol/spot) were incubated with GST–Dab2(1–205) PTB domain and then bound protein visualized with GSH-derivatized HRP. (D) The PTB domain of Dab2 binds to phosphoinositides and FXNPXF internalization motifs synchronously. Thrombin-cleaved Dab2(1–205) (PTB domain) was added to 3 µM, together with 0.4 mg/ml phosphoinositide-containing liposomes alone (lanes a and b) or liposomes plus either 15 µM NWRLKNINSIFDNPVYQKTT (lanes c and d) or NWRLKNINSIFDAPVAQKTT (lanes e and f) peptide and incubated on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. The bound peptide migrates at the dye front and the asterisk denotes the stable ∼25 kDa Dab2 degradation product of the PTB domain.
Fig. 6. The PTB domain of Dab2 binds to polyphosphoinositides. (A) GST–Dab2(1–205) (lanes a–f), GST–epsin 1(1–163) (lanes g and h) or GST (lanes i and j), as indicated, were mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated together on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. (B) Thrombin-cleaved Dab2(1–205) (lanes a–f) or GST (lanes g and h) was mixed with either control (lanes a and b) or phosphoinositide-containing (lanes c–j) liposomes and incubated on ice. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue or transferred to nitrocellulose. The blot was probed with an anti-Dab2 mAb. The asterisk denotes a stable ∼25 kDa thrombin degradation product that binds phosphoinositides and reacts with the anti-Dab2 mAb 52. (C) Nitrocellulose-immobilized phosphoinositides (100, 50, 25, 12.5, 6.2, 3.1, 1.5 pmol/spot) were incubated with GST–Dab2(1–205) PTB domain and then bound protein visualized with GSH-derivatized HRP. (D) The PTB domain of Dab2 binds to phosphoinositides and FXNPXF internalization motifs synchronously. Thrombin-cleaved Dab2(1–205) (PTB domain) was added to 3 µM, together with 0.4 mg/ml phosphoinositide-containing liposomes alone (lanes a and b) or liposomes plus either 15 µM NWRLKNINSIFDNPVYQKTT (lanes c and d) or NWRLKNINSIFDAPVAQKTT (lanes e and f) peptide and incubated on ice for 60 min. After centrifugation, aliquots of 1/25 of each supernatant (S) or 1/4 of each pellet (P) were analyzed by SDS–PAGE and stained with Coomassie Blue. The bound peptide migrates at the dye front and the asterisk denotes the stable ∼25 kDa Dab2 degradation product of the PTB domain.
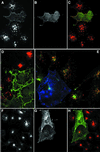
Fig. 7. Dab2 PTB domain overexpression selectively interferes with LDL uptake. COS-7 cells transiently transfected with myc-tagged Dab2 PTBx2 were pulsed for 15 min with 20 µg/ml diI-LDL alone (A–D), both diI-LDL and 25 µg/ml biotinylated transferrin (E) or transferrin alone (F–H) for 15 min prior to fixation. Transfected cells were identified using mAb 9E10 (B–D, G and H, in green in C, D and H) and transferrin with Alexa 488– (E) or Alexa 594–streptavidin (F and H). A single confocal section of triple-labeled cells is shown in (E) (myc epitope, blue; transferrin, green; LDL, red).
Fig. 8. Dab2 co-operates with AP-2 to drive the assembly of invaginated clathrin buds upon lipid membranes. Clathrin was added to 10% PtdIns(4,5)P2-containing lipid monolayers pre-incubated in the absence (A, inset) or presence (A and B) of GST–Dab2(1–368). An EM grid was used to remove each monolayer and then negatively stained. Bar = 50 nm. (C) Phosphoinositide-containing liposomes were first pre-incubated with AP-2 (lanes c, d, g–j and m, n), GST–Dab2(1–368) (lanes e–j) or GST–epsin 1(1–407) (lanes k–n) at 4°C for 60 min as indicated. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/40 of each supernatant (S) and 1/4 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue. Before centrifugation, Triton X-100 (1% final) was added in one reaction (lanes i and j). (D) PtdIns(4,5)P2-containing liposomes were first pre-incubated with (lanes a–j) or without (lanes k and l) thrombin-cleaved Dab2(1–368) at 4°C for 60 min. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with increasing amounts of purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/25 of each supernatant (S) and 1/5 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue.
Fig. 8. Dab2 co-operates with AP-2 to drive the assembly of invaginated clathrin buds upon lipid membranes. Clathrin was added to 10% PtdIns(4,5)P2-containing lipid monolayers pre-incubated in the absence (A, inset) or presence (A and B) of GST–Dab2(1–368). An EM grid was used to remove each monolayer and then negatively stained. Bar = 50 nm. (C) Phosphoinositide-containing liposomes were first pre-incubated with AP-2 (lanes c, d, g–j and m, n), GST–Dab2(1–368) (lanes e–j) or GST–epsin 1(1–407) (lanes k–n) at 4°C for 60 min as indicated. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/40 of each supernatant (S) and 1/4 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue. Before centrifugation, Triton X-100 (1% final) was added in one reaction (lanes i and j). (D) PtdIns(4,5)P2-containing liposomes were first pre-incubated with (lanes a–j) or without (lanes k and l) thrombin-cleaved Dab2(1–368) at 4°C for 60 min. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with increasing amounts of purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/25 of each supernatant (S) and 1/5 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue.
Fig. 8. Dab2 co-operates with AP-2 to drive the assembly of invaginated clathrin buds upon lipid membranes. Clathrin was added to 10% PtdIns(4,5)P2-containing lipid monolayers pre-incubated in the absence (A, inset) or presence (A and B) of GST–Dab2(1–368). An EM grid was used to remove each monolayer and then negatively stained. Bar = 50 nm. (C) Phosphoinositide-containing liposomes were first pre-incubated with AP-2 (lanes c, d, g–j and m, n), GST–Dab2(1–368) (lanes e–j) or GST–epsin 1(1–407) (lanes k–n) at 4°C for 60 min as indicated. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/40 of each supernatant (S) and 1/4 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue. Before centrifugation, Triton X-100 (1% final) was added in one reaction (lanes i and j). (D) PtdIns(4,5)P2-containing liposomes were first pre-incubated with (lanes a–j) or without (lanes k and l) thrombin-cleaved Dab2(1–368) at 4°C for 60 min. After recovery by centrifugation, each liposome pellet was resuspended and then incubated at 4°C for 60 min with increasing amounts of purified clathrin trimers in the presence of carrier BSA. After centrifugation, aliquots of 1/25 of each supernatant (S) and 1/5 of each pellet (P) were resolved by SDS–PAGE and stained with Coomassie Blue.
Similar articles
- Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2.
Morris SM, Cooper JA. Morris SM, et al. Traffic. 2001 Feb;2(2):111-23. doi: 10.1034/j.1600-0854.2001.020206.x. Traffic. 2001. PMID: 11247302 - Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly.
Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Edeling MA, et al. Dev Cell. 2006 Mar;10(3):329-42. doi: 10.1016/j.devcel.2006.01.016. Dev Cell. 2006. PMID: 16516836 - The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery.
Mishra SK, Watkins SC, Traub LM. Mishra SK, et al. Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16099-104. doi: 10.1073/pnas.252630799. Epub 2002 Nov 25. Proc Natl Acad Sci U S A. 2002. PMID: 12451172 Free PMC article. - Epsins: adaptors in endocytosis?
Wendland B. Wendland B. Nat Rev Mol Cell Biol. 2002 Dec;3(12):971-7. doi: 10.1038/nrm970. Nat Rev Mol Cell Biol. 2002. PMID: 12461563 Review. - Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection.
Traub LM. Traub LM. J Cell Biol. 2003 Oct 27;163(2):203-8. doi: 10.1083/jcb.200309175. J Cell Biol. 2003. PMID: 14581447 Free PMC article. Review.
Cited by
- Dab2 gene variant is associated with increased coronary artery disease risk in Chinese Han population.
Wang Y, Wang Y, Adi D, He X, Liu F, Abudesimu A, Fu Z, Ma Y. Wang Y, et al. Medicine (Baltimore). 2020 Jul 2;99(27):e20924. doi: 10.1097/MD.0000000000020924. Medicine (Baltimore). 2020. PMID: 32629690 Free PMC article. - Spatial regulation of VEGF receptor endocytosis in angiogenesis.
Nakayama M, Nakayama A, van Lessen M, Yamamoto H, Hoffmann S, Drexler HC, Itoh N, Hirose T, Breier G, Vestweber D, Cooper JA, Ohno S, Kaibuchi K, Adams RH. Nakayama M, et al. Nat Cell Biol. 2013 Mar;15(3):249-60. doi: 10.1038/ncb2679. Epub 2013 Jan 27. Nat Cell Biol. 2013. PMID: 23354168 Free PMC article. - Disabled-2 heterozygous mice are predisposed to endometrial and ovarian tumorigenesis and exhibit sex-biased embryonic lethality in a p53-null background.
Yang DH, Fazili Z, Smith ER, Cai KQ, Klein-Szanto A, Cohen C, Horowitz IR, Xu XX. Yang DH, et al. Am J Pathol. 2006 Jul;169(1):258-67. doi: 10.2353/ajpath.2006.060036. Am J Pathol. 2006. PMID: 16816378 Free PMC article. - Double-membrane gap junction internalization requires the clathrin-mediated endocytic machinery.
Gumpert AM, Varco JS, Baker SM, Piehl M, Falk MM. Gumpert AM, et al. FEBS Lett. 2008 Aug 20;582(19):2887-92. doi: 10.1016/j.febslet.2008.07.024. Epub 2008 Jul 24. FEBS Lett. 2008. PMID: 18656476 Free PMC article. - The non-canonical roles of clathrin and actin in pathogen internalization, egress and spread.
Humphries AC, Way M. Humphries AC, et al. Nat Rev Microbiol. 2013 Aug;11(8):551-60. doi: 10.1038/nrmicro3072. Nat Rev Microbiol. 2013. PMID: 24020073 Review.
References
- Arneson L.S., Kunz,J., Anderson,R.A. and Traub,L.M. (1999) Coupled inositide phosphorylation and phospholipase D activation initiates clathrin-coat assembly on lysosomes. J. Biol. Chem., 274, 17794–17805. - PubMed
- Brett T.J., Traub,L.M. and Fremont,D.H. (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure, 10, 797–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials