Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2 - PubMed (original) (raw)
Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2
Olivier Stevaux et al. EMBO J. 2002.
Abstract
RBF1, a Drosophila pRB family homolog, is required for cell cycle arrest and the regulation of E2F-dependent transcription. Here, we describe the properties of RBF2, a second family member. RBF2 represses E2F transcription and is present at E2F-regulated promoters. Analysis of in vivo protein complexes reveals that RBF1 and RBF2 interact with different subsets of E2F proteins. dE2F1, a potent transcriptional activator, is regulated specifically by RBF1. In contrast, RBF2 binds exclusively to dE2F2, a form of E2F that functions as a transcriptional repressor. We find that RBF2-mediated repression requires dE2F2. More over, RBF2 and dE2F2 act synergistically to antagonize dE2F1-mediated activation, and they co-operate to block S phase progression in transgenic animals. The network of interactions between RBF1 or RBF2 and dE2F1 or dE2F2 reveals how the activities of these proteins are integrated. These results suggest that there is a remarkable degree of symmetry in the arrangement of E2F and RB family members in mammalian cells and in DROSOPHILA.
Figures
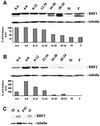
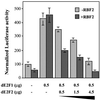
Fig. 1. RBF2 is an RBF1 homolog more closely related to p107 than to pRB. (A) Alignment of RBF1 and RBF2. Conserved domains between RBF2 and other pocket proteins are underlined: the A and B halves of the pocket, multiple cyclin/CDK phosphorylation sites (*), and the N-terminal (N) and spacer (S) domains that are conserved in p107 and p130, but not in pRB. (B) Alignment between the N-terminal domain of RBF2 and conserved regions of p107 and p130. (C) Alignment between the spacer sequence of RBF2 and portions of the p107 and p130 spacer sequences. (D) Phylogenetic tree of RB-related proteins. The percentages of identity between each pocket protein and RBF2 are shown in parenthesis.
Fig. 2. RBF1 and RBF2 are differentially expressed throughout development. W1118 embryos were collected at intervals of 4 h. Six collections covered the 24 h of embryogenesis at 25°C. Protein extracts from the staged embryos and from male or female adults were analyzed by western blotting with monoclonal antibodies specific for (A) RBF1 or (B) RBF2 and with a tubulin antibody for loading control. The charts show the levels of the RBF1 and RBF2 as a percentage of the level in the earliest extract, normalized for tubulin levels. (C) Analysis of protein extracts, corresponding to one adult male (M), one adult female (F), one adult female after resection of ovaries (F\O), and dissected ovaries from one female (O), by western blotting with an RBF2 or a tubulin antibody.
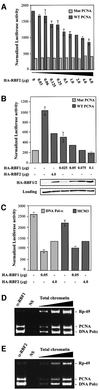
Fig. 3. RBF1 and RBF2 both repress E2F transcription. (A) SL2 cells were transfected with wild-type or mutant PCNA reporters and with various amounts of PIE4-HARBF2. (B) SL2 cells were transfected with the PCNA reporter and with 4 µg of PIE4-HARBF2 and various amounts of PIE4-HARBF1. An HA antibody was used to monitor the expression of HA-RBF1 and HA-RBF2 in transfected cells. (C) A similar experiment was conducted using the _DNA Pol_α (light bars) and the MCM3 (dark bars) reporter constructs. (D and E) RBF1 and RBF2 occupy E2F-regulated promoters in vivo. Chromatin samples precipitated with RBF1, RBF2 or non-specific (NS) antibodies were used as templates for the amplification by PCR of the PCNA, the _DNA Pol_α and the control RP49 promoter regions with specific primers. Increasing amounts of total chromatin were used to ensure the linearity of the PCR amplification.
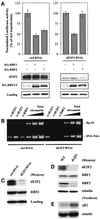
Fig. 4. RBF2 does not inactivate dE2F1-induced transcription. PIE4-mycdE2F1 (0.5 µg) was transfected with the PCNA reporter and various amounts of PIE4-HARBF1 or PIE4-HARBF2. Clear bars are transfected with the mutant PCNA reporter, dark bars are transfected with the wild-type construct. In the absence of PIE4-mycdE2F1, the activation of the wild-type PCNA promoter is due to the endogenous E2F activity in SL2 cells.
Fig. 5. Protein interactions between RBF and dE2F proteins. (A) RBF2 co-immunoprecipitates with dE2F2 but not with dE2F1. α-dDP, α-dE2F1, α-dE2F2 or control (α-E1A) antibodies were used to prepare immune complexes from SL2 cell extracts (+). Western blot analysis was performed using an α-RBF2-specific monoclonal antibody. After stripping, the membrane was re-probed with an α-RBF1-specific monoclonal antibody. (–) represents control immunoprecipitations using lysis buffer. (B) dE2F1 co-immunoprecipitates with RBF1 but not with RBF2. Immune complexes were prepared from SL2 cell extracts (+) using antibodies specific for RBF1, RBF2, dE2F1 or a control (E1A). The western blot was probed with a guinea pig α-dE2F1 antibody, then stripped and re-probed with an α-RBF2 antibody. This immunoprecipitation was subsequently repeated and the western blot was probed with an α-dE2F2 rabbit polyclonal antibody. (C) Summary of the interaction pattern between dE2Fs and RBFs.
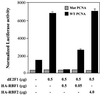
Fig. 6. dE2F2 is required for RBF2 repression, localization, and expression. (A) SL2 cells were treated with white (ctrl) or de2f2 dsRNA and transfected with the MCM3 reporter together with either 0.05 µg of PIE4-HARBF1 or 4 µg of PIE4-HARBF2. Depletion of dE2F2 increases transcription from the PCNA reporter by 35%. To compare the ability of RBF1 and RBF2 to repress in these different conditions, the levels of luciferase activity are expressed as a percentage of the luciferase activity observed in the absence of any RBF expression plasmid. Western blot analysis with an α-dE2F2 or an α-HA antibody confirms the specific depletion of dE2F2, and the expression of equivalent levels of HARBF1 and HARBF2. (B) RBF2 occupancy of the _DNA Pol_α promoter requires dE2F2 in vivo. Chromatin samples precipitated with dE2F1, dE2F2, or RBF2 antibodies were used as templates for the amplification by PCR of the _DNA Pol_α, and the control RP49 promoter regions with specific primers. (C) RBF2 levels are only slightly reduced in cells treated with dE2F2 RNAi. (D) dE2F2 is required for the maintenance of RBF2 protein levels in vivo. Western blotting analysis of protein extracts from wild-type or de2f2 mutants larvae using α-dE2F2, α-RBF1, α-RBF2 or α-tubulin antibodies. (E) The loss of the RBF2 protein in de2f2 mutants is not due to transcriptional changes. Northern blot analysis of total RNA from wild-type or de2f2 mutant larvae using an rbf2 or a tubulin radiolabeled antisense riboprobe.
Fig. 7. RBF2 and dE2F2 cooperate to antagonize dE2F1-induced transcription. SL2 cells were transfected with the PCNA reporter, 0.5 µg of PIE4-mycdE2F1, and increasing amounts of PIE4-mycdE2F2. E2F-dependent transcription was detected in either the presence (4 µg of PIE4-HARBF2; dark gray bars) or absence (4 µg of empty PIE4; light gray bars) of exogenous RBF2.
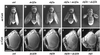
Fig. 8. RBF2 and dE2F2 transgenes have a synergistic effect in vivo. Scan electron microscopy (SEM) of transgenic flies of the following genotypes: (A) _ap-Gal4/+;_UAS-lacZ/+; (B) _ap-Gal4/+;_UAS-de2f2a/+; (C) _ap-Gal4/_UAS-rbf2a; (D) _ap-Gal4/_UAS-_rbf2a;_UAS-de2f2a/+; (E) _ap-Gal4/+;_UAS-_de2f2b-_UAS-rbf2b/+; (F) _ey-Gal4/+;_UAS-lacZ/+; (G) _ey-Gal4/+;_UAS-de2f2b/+; (H) _ey-Gal4/+;_UAS-rbf2b/+; (I) ey-Gal4/+;UAS-de2f2b- UAS-rbf2b/+; (J) _ey-Gal4/_UAS-rbf1. SEM (A–E), 5 kV, 32×; SEM (F–J), 10 kV, 200× (Scale bars = 1 mm).
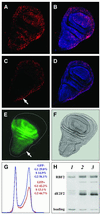
Fig. 9. Overexpression of RBF2 and dE2F2 blocks S phase entry and causes cells to accumulate in the G1 phase of the cell cycle. BrdU incorporation in third instar larval wing discs of the following genotype: (A and B) _ap-_Gal4/+;UAS-lacZ/+; (C and D) _ap-_Gal4/UAS- _rbf2a;_UAS-de2f2a/+. Red, anti-BrdU staining; blue, yo-yo DNA staining. (E) EGFP pattern of expression in an _ap-_Gal4/+;UAS-EGFP/+ third instar larval wing disc. (F) Nomarski imaging of the same disc as in (E). (G) FACS profile of the GFP-negative wild-type cells (blue graph) as compared with GFP-positive cells overexpressing dE2F2 and RBF2 (red graph). (H) Western blot analysis with RBF2 or dE2F2 antibodies of protein extracts from ten third instar larval wing disc of the following genotypes: _ap-_Gal4/+;UAS-lacZ/+ (lane 1); _ap-_Gal4/UAS-_rbf2a;_UAS-de2f2a/+ (lane 2); _ap-_Gal4/+;UAS-de2f2b-UAS-rbf2b/+ (lane 3).
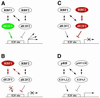
Fig. 10. Model of E2F transcriptional regulation by pocket proteins in Drosophila. The transcriptional output from known E2F-regulated promoters depends on the relative activity of activator and repressor complexes. Activation is depicted in green, repression in red. (A) The expression of dE2F1, or its release from RBF1, strongly induces transcription. (B) RBF1 binds to the activation domain of dE2F1 and inactivates dE2F1-induced transcription. RBF1 can also interact with dE2F2, but this interaction is not required for RBF1 to repress E2F-dependent promoters. (C) RBF2, however, does not associate with dE2F1. RBF2 forms complexes with dE2F2 that antagonize dE2F1- induced transcription indirectly and causes a shift towards repression. (D) The pattern of dE2F–RBF interactions in Drosophila parallels the arrangement of the mammalian E2F/pocket protein pathway.
Similar articles
- Genome-Wide Analysis of Drosophila RBf2 Protein Highlights the Diversity of RB Family Targets and Possible Role in Regulation of Ribosome Biosynthesis.
Wei Y, Mondal SS, Mouawad R, Wilczyński B, Henry RW, Arnosti DN. Wei Y, et al. G3 (Bethesda). 2015 May 20;5(7):1503-15. doi: 10.1534/g3.115.019166. G3 (Bethesda). 2015. PMID: 25999584 Free PMC article. - p55, the Drosophila ortholog of RbAp46/RbAp48, is required for the repression of dE2F2/RBF-regulated genes.
Taylor-Harding B, Binné UK, Korenjak M, Brehm A, Dyson NJ. Taylor-Harding B, et al. Mol Cell Biol. 2004 Oct;24(20):9124-36. doi: 10.1128/MCB.24.20.9124-9136.2004. Mol Cell Biol. 2004. PMID: 15456884 Free PMC article. - RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes.
Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. Korenjak M, et al. Mol Cell Biol. 2012 Nov;32(21):4375-87. doi: 10.1128/MCB.00536-12. Epub 2012 Aug 27. Mol Cell Biol. 2012. PMID: 22927638 Free PMC article. - Molecular mechanisms of E2F-dependent activation and pRB-mediated repression.
Frolov MV, Dyson NJ. Frolov MV, et al. J Cell Sci. 2004 May 1;117(Pt 11):2173-81. doi: 10.1242/jcs.01227. J Cell Sci. 2004. PMID: 15126619 Review. - The E2F family: specific functions and overlapping interests.
Attwooll C, Lazzerini Denchi E, Helin K. Attwooll C, et al. EMBO J. 2004 Dec 8;23(24):4709-16. doi: 10.1038/sj.emboj.7600481. Epub 2004 Nov 11. EMBO J. 2004. PMID: 15538380 Free PMC article. Review.
Cited by
- Novel functions for the transcription factor E2F4 in development and disease.
Hsu J, Sage J. Hsu J, et al. Cell Cycle. 2016 Dec;15(23):3183-3190. doi: 10.1080/15384101.2016.1234551. Epub 2016 Oct 18. Cell Cycle. 2016. PMID: 27753528 Free PMC article. Review. - Cross-species identification of PIP5K1-, splicing- and ubiquitin-related pathways as potential targets for RB1-deficient cells.
Parkhitko AA, Singh A, Hsieh S, Hu Y, Binari R, Lord CJ, Hannenhalli S, Ryan CJ, Perrimon N. Parkhitko AA, et al. PLoS Genet. 2021 Feb 16;17(2):e1009354. doi: 10.1371/journal.pgen.1009354. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33591981 Free PMC article. - Paradoxical instability-activity relationship defines a novel regulatory pathway for retinoblastoma proteins.
Acharya P, Raj N, Buckley MS, Zhang L, Duperon S, Williams G, Henry RW, Arnosti DN. Acharya P, et al. Mol Biol Cell. 2010 Nov 15;21(22):3890-901. doi: 10.1091/mbc.E10-06-0520. Epub 2010 Sep 22. Mol Biol Cell. 2010. PMID: 20861300 Free PMC article. - RBF and Rno promote photoreceptor differentiation onset through modulating EGFR signaling in the Drosophila developing eye.
Sukhanova MJ, Steele LJ, Zhang T, Gordon GM, Du W. Sukhanova MJ, et al. Dev Biol. 2011 Nov 15;359(2):190-8. doi: 10.1016/j.ydbio.2011.08.018. Epub 2011 Sep 2. Dev Biol. 2011. PMID: 21920355 Free PMC article. - A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development.
Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Näär AM. Mulligan P, et al. Mol Cell. 2011 Jun 10;42(5):689-99. doi: 10.1016/j.molcel.2011.04.020. Epub 2011 May 19. Mol Cell. 2011. PMID: 21596603 Free PMC article.
References
- Asano M., Nevins,J.R. and Wharton,R.P.E. (1996) Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev., 10, 1422–1432. - PubMed
- Benvenuto G. et al. (2000) The tuberous sclerosis (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene, 19, 6306–6316. - PubMed
- Bosco G., Du,W. and Orr-Weaver,T. (2001) DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol., 3, 289–295. - PubMed
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. - PubMed
- Brehm A. and Kouzarides,T. (1999) Retinoblastoma protein meets chromatin. Trends Biochem. Sci., 24, 142–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA88474/CA/NCI NIH HHS/United States
- R01 GM053203/GM/NIGMS NIH HHS/United States
- F32 CA93045/CA/NCI NIH HHS/United States
- F32 CA093045/CA/NCI NIH HHS/United States
- F32 CA088474/CA/NCI NIH HHS/United States
- GM53203/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases