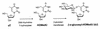Expression of the human DNA glycosylase hSMUG1 in Trypanosoma brucei causes DNA damage and interferes with J biosynthesis - PubMed (original) (raw)
Expression of the human DNA glycosylase hSMUG1 in Trypanosoma brucei causes DNA damage and interferes with J biosynthesis
Sebastian Ulbert et al. Nucleic Acids Res. 2002.
Abstract
In kinetoplastid flagellates such as Trypanosoma brucei, a small percentage of the thymine residues in the nuclear DNA is replaced by the modified base beta-D-glucosyl-hydroxymethyluracil (J), mostly in repetitive sequences like the telomeric GGGTTA repeats. In addition, traces of 5-hydroxymethyluracil (HOMeUra) are present. Previous work has suggested that J is synthesised in two steps via HOMedU as an intermediate, but as J synthesising enzymes have not yet been identified, the biosynthetic pathway remains unclear. To test a model in which HOMeUra functions as a precursor of J, we introduced an inducible gene for the human DNA glycosylase hSMUG1 into bloodstream form T.brucei. In higher eukaryotes SMUG1 excises HOMeUra as part of the base excision repair system. We show that expression of the gene in T.brucei leads to massive DNA damage in J-modified sequences and results in cell cycle arrest and, eventually, death. hSMUG1 also reduces the J content of the trypanosome DNA. This work supports the idea that HOMeUra is a precursor of J, freely accessible to a DNA glycosylase.
Figures
Figure 1
Speculative two-step model for J biosynthesis. At the DNA level, thymidine is first converted to HOMedU by a putative thymidine 7-hydroxylase and then HOMedU is converted into J by a putative β-glucosyltransferase. From van Leeuwen et al. (7).
Figure 2
Schematic drawing of the construct pHDhSMUG1 used to yield inducible expression of hSMUG1. The construct is based on pHD615 (16). Ti-P, tetracycline-inducible promoter; V, constitutive VSG promoter; hSMUG1, human single-strand-selective monofunctional uracil-DNA glycosylase; PAC, puromycin acetyltransferase; black boxes, trypanosome-specific RNA processing signals (for details see 16); hatched boxes, flanks for integration into the non-transcribed rRNA spacer in opposite orientation to the direction of endogenous transcription. The solid black line represents pGEM vector sequence. The two open circles are tetracycline repressor molecules derived from transcription of the repressor gene in the α/β-tubulin arrays (data not shown). Binding of the repressor is inhibited by tetracycline.
Figure 3
(A) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing HOMedU paired with G (the HOMedU:A base pair yielded similar results; not shown). The substrate band represents the full-length oligonucleotide, the product band is the cut oligonucleotide after excision of the HOMeUra residue. The lane on the right (–) is the substrate without lysate. (B) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing uracil paired with G. Lane 1, substrate without lysate; lane 2, wild-type cells; lane 3, hSMUG1 transfectants with tetracycline; lane 4, wild-type cells with UGI; lane 5, hSMUG1 transfectants with tetracycline and UGI. (C) Western blot analysis of the hSMUG1 transfectants. The blot was incubated with an anti-HA tag antibody, stripped, and incubated with an anti-aldolase antibody as a loading control. 3 × 106 cell equivalents were loaded per lane.
Figure 3
(A) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing HOMedU paired with G (the HOMedU:A base pair yielded similar results; not shown). The substrate band represents the full-length oligonucleotide, the product band is the cut oligonucleotide after excision of the HOMeUra residue. The lane on the right (–) is the substrate without lysate. (B) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing uracil paired with G. Lane 1, substrate without lysate; lane 2, wild-type cells; lane 3, hSMUG1 transfectants with tetracycline; lane 4, wild-type cells with UGI; lane 5, hSMUG1 transfectants with tetracycline and UGI. (C) Western blot analysis of the hSMUG1 transfectants. The blot was incubated with an anti-HA tag antibody, stripped, and incubated with an anti-aldolase antibody as a loading control. 3 × 106 cell equivalents were loaded per lane.
Figure 3
(A) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing HOMedU paired with G (the HOMedU:A base pair yielded similar results; not shown). The substrate band represents the full-length oligonucleotide, the product band is the cut oligonucleotide after excision of the HOMeUra residue. The lane on the right (–) is the substrate without lysate. (B) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing uracil paired with G. Lane 1, substrate without lysate; lane 2, wild-type cells; lane 3, hSMUG1 transfectants with tetracycline; lane 4, wild-type cells with UGI; lane 5, hSMUG1 transfectants with tetracycline and UGI. (C) Western blot analysis of the hSMUG1 transfectants. The blot was incubated with an anti-HA tag antibody, stripped, and incubated with an anti-aldolase antibody as a loading control. 3 × 106 cell equivalents were loaded per lane.
Figure 4
Growth curve of hSMUG1 transfectants with (triangles) and without (squares) tetracycline (1 µg/ml).
Figure 5
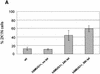
(A) Percentage of cells with two kinetoplasts and one nucleus in cultures of trypanosomes exposed to various levels of endogenous hSMUG1. (B) Measurement of the DNA content of trypanosomes using flow cytometry. (1) Wild-type cells; (2) hSMUG1 transfectants after 40 h in tetracycline (1 µg/ml). Sub-G1 cells are marked with an asterisk. (C and D) Effect of nucleoside feeding on hSMUG1 induction. (C) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with HOMedU (200 µM for 48 h). Dotted lines are non-fed control cells. (D) Growth curve of _hSMUG_1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with BrdU (100 µM for 72 h). Dotted lines are non-fed control cells.
Figure 5
(A) Percentage of cells with two kinetoplasts and one nucleus in cultures of trypanosomes exposed to various levels of endogenous hSMUG1. (B) Measurement of the DNA content of trypanosomes using flow cytometry. (1) Wild-type cells; (2) hSMUG1 transfectants after 40 h in tetracycline (1 µg/ml). Sub-G1 cells are marked with an asterisk. (C and D) Effect of nucleoside feeding on hSMUG1 induction. (C) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with HOMedU (200 µM for 48 h). Dotted lines are non-fed control cells. (D) Growth curve of _hSMUG_1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with BrdU (100 µM for 72 h). Dotted lines are non-fed control cells.
Figure 5
(A) Percentage of cells with two kinetoplasts and one nucleus in cultures of trypanosomes exposed to various levels of endogenous hSMUG1. (B) Measurement of the DNA content of trypanosomes using flow cytometry. (1) Wild-type cells; (2) hSMUG1 transfectants after 40 h in tetracycline (1 µg/ml). Sub-G1 cells are marked with an asterisk. (C and D) Effect of nucleoside feeding on hSMUG1 induction. (C) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with HOMedU (200 µM for 48 h). Dotted lines are non-fed control cells. (D) Growth curve of _hSMUG_1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with BrdU (100 µM for 72 h). Dotted lines are non-fed control cells.
Figure 5
(A) Percentage of cells with two kinetoplasts and one nucleus in cultures of trypanosomes exposed to various levels of endogenous hSMUG1. (B) Measurement of the DNA content of trypanosomes using flow cytometry. (1) Wild-type cells; (2) hSMUG1 transfectants after 40 h in tetracycline (1 µg/ml). Sub-G1 cells are marked with an asterisk. (C and D) Effect of nucleoside feeding on hSMUG1 induction. (C) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with HOMedU (200 µM for 48 h). Dotted lines are non-fed control cells. (D) Growth curve of _hSMUG_1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with BrdU (100 µM for 72 h). Dotted lines are non-fed control cells.
Figure 6
Agarose gel electrophoresis on wild-type cells and hSMUG1 transfectants cultured with and without tetracycline. Whole cells were cast in agarose blocks, digested with proteinase K and subjected to electrophoresis. The gel was run at a low voltage, blotted and hybridised with a telomeric repeat probe. Subsequently, the membrane was stripped and hybridised with a probe for the α/β-tubulin gene array.
Figure 7
J measurement on genomic DNA of wild-type trypanosomes and hSMUG1 cells cultured with and without tetracycline. Aliquots of 50 ng of DNA were diluted in 1:1 steps and spotted onto a membrane which was then incubated with an anti-J antibody. The membrane was stripped and hybridised with a probe for the α/β-tubulin gene array to calculate the difference in J level corrected for DNA loading (data not shown).
Similar articles
- Base J, found in nuclear DNA of Trypanosoma brucei, is not a target for DNA glycosylases.
Ulbert S, Eide L, Seeberg E, Borst P. Ulbert S, et al. DNA Repair (Amst). 2004 Feb 3;3(2):145-54. doi: 10.1016/j.dnarep.2003.10.009. DNA Repair (Amst). 2004. PMID: 14706348 - Base J: discovery, biosynthesis, and possible functions.
Borst P, Sabatini R. Borst P, et al. Annu Rev Microbiol. 2008;62:235-51. doi: 10.1146/annurev.micro.62.081307.162750. Annu Rev Microbiol. 2008. PMID: 18729733 Review. - Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei.
van Leeuwen F, Kieft R, Cross M, Borst P. van Leeuwen F, et al. Mol Cell Biol. 1998 Oct;18(10):5643-51. doi: 10.1128/MCB.18.10.5643. Mol Cell Biol. 1998. PMID: 9742081 Free PMC article. - The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands.
van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. van Leeuwen F, et al. Nucleic Acids Res. 1996 Jul 1;24(13):2476-82. doi: 10.1093/nar/24.13.2476. Nucleic Acids Res. 1996. PMID: 8692684 Free PMC article. - beta-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida.
Borst P, van Leeuwen F. Borst P, et al. Mol Biochem Parasitol. 1997 Dec 1;90(1):1-8. doi: 10.1016/s0166-6851(97)00170-9. Mol Biochem Parasitol. 1997. PMID: 9497027 Review.
Cited by
- The DNA damage response is developmentally regulated in the African trypanosome.
Vieira-da-Rocha JP, Passos-Silva DG, Mendes IC, Rocha EA, Gomes DA, Machado CR, McCulloch R. Vieira-da-Rocha JP, et al. DNA Repair (Amst). 2019 Jan;73:78-90. doi: 10.1016/j.dnarep.2018.11.005. Epub 2018 Nov 14. DNA Repair (Amst). 2019. PMID: 30470509 Free PMC article. - Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing.
Genest PA, Baugh L, Taipale A, Zhao W, Jan S, van Luenen HG, Korlach J, Clark T, Luong K, Boitano M, Turner S, Myler PJ, Borst P. Genest PA, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2102-15. doi: 10.1093/nar/gkv095. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662217 Free PMC article. - Identification of the glucosyltransferase that converts hydroxymethyluracil to base J in the trypanosomatid genome.
Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. Bullard W, et al. J Biol Chem. 2014 Jul 18;289(29):20273-82. doi: 10.1074/jbc.M114.579821. Epub 2014 Jun 2. J Biol Chem. 2014. PMID: 24891501 Free PMC article. - Trypanosoma brucei brucei: thymine 7-hydroxylase-like proteins.
Simmons JM, Koslowsky DJ, Hausinger RP. Simmons JM, et al. Exp Parasitol. 2010 Apr;124(4):453-8. doi: 10.1016/j.exppara.2009.11.012. Epub 2009 Nov 27. Exp Parasitol. 2010. PMID: 19945457 Free PMC article. - Fe(II)/alpha-ketoglutarate hydroxylases involved in nucleobase, nucleoside, nucleotide, and chromatin metabolism.
Simmons JM, Müller TA, Hausinger RP. Simmons JM, et al. Dalton Trans. 2008 Oct 14;(38):5132-42. doi: 10.1039/b803512a. Epub 2008 Jun 27. Dalton Trans. 2008. PMID: 18813363 Free PMC article.
References
- Gommers-Ampt J., van Leeuwen,F., de Beer,A.L.J., Vliegenthart,J.F.G., Didzdaroglu,M., Kowalak,J.A. and Borst,P. (1993) Beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan Trypanosoma brucei. Cell, 75, 1129–1136. - PubMed
- van Leeuwen F., Kieft,R., Cross,M. and Borst,P. (2000) Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol., 109, 133–145. - PubMed