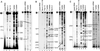Beta -Globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon - PubMed (original) (raw)
Beta -Globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon
Audrey Stevens et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Previous work showed that human beta-globin mRNAs harboring a premature termination codon are degraded in the erythroid tissues of mice to products that lack sequences from the mRNA 5' end but contain a 5' cap-like structure. Whether these decay products are the consequence of endonucleolytic or 5'-to-3' exonucleolytic activity is unclear. We report that this beta-globin mRNA decay pathway is recapitulated in cultured mouse erythroleukemia (MEL) cells and targets nonsense-free mRNA to a lesser extent than nonsense-containing mRNA. S1 nuclease mapping and primer extension demonstrated that 70-80% of decay product 5' ends contain a UG dinucleotide. Detection of upstream counterparts of these decay products indicates that they are generated by endonucleolytic activity. Both crude and partially purified polysome extracts prepared from MEL cells contain an endonucleolytic activity that generates decay products comparable to those observed in vivo. These data suggest that an endonuclease with preference for UG dinucleotides is involved in the degradation of nonsense-containing and, to a lesser extent, nonsense-free human beta-globin mRNAs in mouse erythroid cells.
Figures
Figure 1
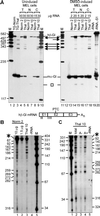
S1 nuclease mapping of 3′ decay products of normal and nonsense-containing human β-globin transcripts. (A) Mice transgenic for a nonsense-free (Norm) or nonsense-containing (Thal) human β-globin allele were made anemic, and RNA was isolated from peripheral blood (13). Additionally, MEL cells stably expressing the nonsense-free (Norm 2) or nonsense-containing (Thal 10) human β-globin allele were propagated in the absence (uninduced) or presence (induced) of 2% DMSO, and RNA was isolated from total (T), nuclear (N), and cytoplasmic (C) fractions (19). A 5′-32P-labeled 590-bp _Mse_I–_Mse_I fragment of human β-globin cDNA and a 5′-32P-labeled 183-bp _Bam_HI–_Bam_HI fragment of mouse α-globin cDNA were used as probes in S1 nuclease mapping (13). Full-length human β-globin mRNA protected 524 nt (hβ-Gl mRNA), mouse α-globin mRNA protected 183 nt (mα-Gl mRNA), and mRNA decay products are identified by arrows. The 159-nt protected fragment (□) derives from intron 2-containing human β-globin pre-mRNA (data not shown). Total (T), nuclear (N), and cytoplasmic (C) RNA from each transfectant was assayed by using the amounts specified above each lane (2, 15, or 30 μg). −RNA, no RNA; −S1, no S1 nuclease. The locations of the premature termination codon (PTC) and 5′ decay products are diagrammed. (B and C) A 5′-32P-labeled _Nae_I–_Bam_HI fragment of a human β-globin cDNA was used as probe for S1 nuclease mapping. Full-length mRNA protected 351 nt (✶). In B, 10 μg of total RNA was used except for lane 2, where 1.5 μg was used. In C, 10 μg of total, 10 μg of poly(A)−, and 0.5 μg of poly(A)+ RNA were used. The sizes of the protected fragments are indicated according to the distance from the 5′ end of full-length β-globin mRNA to the left in B and right in C. M, molecular weight standards.
Figure 2
Sequence of human β-globin mRNA, where nucleotides in bold are complementary to DNA primers used in primer extensions.
Figure 3
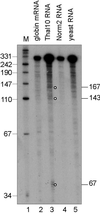
S1 nuclease mapping of 5′ decay products of normal and nonsense-containing human β-globin transcripts. S1 nuclease protection was performed as in Fig. 1 by using 0.74 ng of synthetic β-globin mRNA (lane 2) (21), 15 μg of poly(A)− MEL cell RNA, or 15 μg of yeast RNA and a 3′-labeled _Bfa_I–_Bam_HI probe that spans nucleotides 31–351 of human β-globin mRNA. Decay intermediates are identified with circles.
Figure 4
Primer extension mapping the 5′ ends of the human β-globin mRNA decay products. Total RNA (10 μg), or synthetic human β-globin mRNA (7.2 ng) (21) was annealed to oligonucleotide DNAs (see Fig. 2 and Table 1) complementary to mRNA nucleotides 112–136 (A), 226–250 (B), or 341–365 (C) and assayed by primer extension. Sequencing ladders (T or G ladders) provided molecular size markers, and a control primer extension (synthetic globin RNA) identified sites of reverse transcriptase stalling. Numbers to the left map potential cleavage sites relative to the full-length 5′ end, which is defined as 1. Notably, because Thal 10 mRNA contains one less nucleotide at codon 44 relative to Norm 2 mRNA, the extension band resulting from cleavage at nucleotide 167 is shorter by one nucleotide. Results shown are representative of at least two independently performed experiments that used RNA from transfectants grown and harvested on different days. Full-length products are identified with an asterisk (✶).
Figure 5
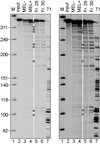
β-globin mRNA endonuclease activity associated with MEL-cell polysomes. (Left) An _in vitro_-synthesized, 5′-32P-labeled transcript consisting of the 5′-most 347 nt of human β-globin mRNA (plus 20 nt of vector) was incubated for 30 min at 37°C with no added protein (lane 2, input), polysome extract from uninduced (MEL−, lane 3) or 48-hr DMSO-induced (MEL+, lane 4) MEL cells, or two fractions containing β-globin mRNA endonuclease activity recovered from Mono S fractionation of the polysome extract (lanes 5 and 6). Lane 7 shows a partial T1 digest of the β-globin transcript; an open circle (○) shows products with counterparts in vivo. Right is a longer exposure of Left.
Similar articles
- An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human beta-globin mRNA in erythroid cells.
Bremer KA, Stevens A, Schoenberg DR. Bremer KA, et al. RNA. 2003 Sep;9(9):1157-67. doi: 10.1261/rna.5720303. RNA. 2003. PMID: 12923263 Free PMC article. - Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products.
Lim SK, Sigmund CD, Gross KW, Maquat LE. Lim SK, et al. Mol Cell Biol. 1992 Mar;12(3):1149-61. doi: 10.1128/mcb.12.3.1149-1161.1992. Mol Cell Biol. 1992. PMID: 1545796 Free PMC article. - Nonsense mutations in the human beta-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation.
Romão L, Inácio A, Santos S, Avila M, Faustino P, Pacheco P, Lavinha J. Romão L, et al. Blood. 2000 Oct 15;96(8):2895-901. Blood. 2000. PMID: 11023527 - Globin mRNA in Friend cells: its structure, function and synthesis.
Curtis PJ. Curtis PJ. Biochim Biophys Acta. 1980 Sep 22;605(3):347-64. doi: 10.1016/0304-419x(80)90016-5. Biochim Biophys Acta. 1980. PMID: 6996728 Review. - Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells.
Jacobson A, Peltz SW. Jacobson A, et al. Annu Rev Biochem. 1996;65:693-739. doi: 10.1146/annurev.bi.65.070196.003401. Annu Rev Biochem. 1996. PMID: 8811193 Review.
Cited by
- RNA degradome analysis reveals DNE1 endoribonuclease is required for the turnover of diverse mRNA substrates in Arabidopsis.
Nagarajan VK, Stuart CJ, DiBattista AT, Accerbi M, Caplan JL, Green PJ. Nagarajan VK, et al. Plant Cell. 2023 May 29;35(6):1936-1955. doi: 10.1093/plcell/koad085. Plant Cell. 2023. PMID: 37070465 Free PMC article. - Approaches for studying PMR1 endonuclease-mediated mRNA decay.
Otsuka Y, Schoenberg DR. Otsuka Y, et al. Methods Enzymol. 2008;448:241-63. doi: 10.1016/S0076-6879(08)02613-X. Methods Enzymol. 2008. PMID: 19111180 Free PMC article. - A recap of RNA recapping.
Trotman JB, Schoenberg DR. Trotman JB, et al. Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1504. doi: 10.1002/wrna.1504. Epub 2018 Sep 5. Wiley Interdiscip Rev RNA. 2019. PMID: 30252202 Free PMC article. Review. - An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human beta-globin mRNA in erythroid cells.
Bremer KA, Stevens A, Schoenberg DR. Bremer KA, et al. RNA. 2003 Sep;9(9):1157-67. doi: 10.1261/rna.5720303. RNA. 2003. PMID: 12923263 Free PMC article. - SMG6 cleavage generates metastable decay intermediates from nonsense-containing β-globin mRNA.
Mascarenhas R, Dougherty JA, Schoenberg DR. Mascarenhas R, et al. PLoS One. 2013 Sep 25;8(9):e74791. doi: 10.1371/journal.pone.0074791. eCollection 2013. PLoS One. 2013. PMID: 24086375 Free PMC article.
References
- Tharun S, Parker R. In: mRNA Metabolism and Post-Transcriptional Gene Regulation. Harford J, Morris D R, editors. New York: Wiley; 1997. pp. 181–200.
- Schoenberg D R, Chernokalskaya E. In: mRNA Metabolism and Post-Transcriptional Gene Regulation. Harford J, Morris D R, editors. New York: Wiley; 1997. pp. 217–240.
- Binder R, Hwang S P, Ratnasabapathy R, Williams D L. J Biol Chem. 1989;264:16910–16918. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM038277/GM/NIGMS NIH HHS/United States
- R01 GM055407/GM/NIGMS NIH HHS/United States
- GM59614/GM/NIGMS NIH HHS/United States
- GM38277/GM/NIGMS NIH HHS/United States
- R01 GM059614/GM/NIGMS NIH HHS/United States
- GM55407/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous