Genetic diversity and selection in the maize starch pathway - PubMed (original) (raw)
Genetic diversity and selection in the maize starch pathway
Sherry R Whitt et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Maize is both phenotypically and genetically diverse. Sequence studies generally confirm the extensive genetic variability in modern maize is consistent with a lack of selection. For more than 6,000 years, Native Americans and modern breeders have exploited the tremendous genetic diversity of maize (Zea mays ssp. mays) to create the highest yielding grain crop in the world. Nonetheless, some loci have relatively low levels of genetic variation, particularly loci that have been the target of artificial selection, like c1 and tb1. However, there is limited information on how selection may affect an agronomically important pathway for any crop. These pathways may retain the signature of artificial selection and may lack genetic variation in contrast to the rest of the genome. To evaluate the impact of selection across an agronomically important pathway, we surveyed nucleotide diversity at six major genes involved in starch metabolism and found unusually low genetic diversity and strong evidence of selection. Low diversity in these critical genes suggests that a paradigm shift may be required for future maize breeding. Rather than relying solely on the diversity within maize or on transgenics, future maize breeding would perhaps benefit from the incorporation of alleles from maize's wild relatives.
Figures
Figure 1
A simplified pathway of starch production in maize and the position of the six sampled genes in the pathway. *, loci with strong evidence of selection (either HKA or Tajima's D test).
Figure 2
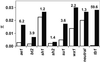
Comparison of silent diversity in maize (white bars) and its wild relative Z. mays ssp. parviglumis (black bars). For each locus, 500–2,700 silent bases were sampled. The numbers above the bars indicate the fold reduction in diversity between maize and Z. m. ssp. parviglumis. Neutral refers to the average of six nonselected genes (adh1, adh2, glb1, hm1, hm2, and te1); tb1 is the only cloned domestication gene from maize (only the highly selected promoter region is shown).
Figure 3
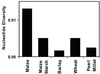
Comparison of nucleotide diversity in maize and various grass crops. Maize data are the average diversity at random loci from Tenaillon et al. (2), the maize starch average is from the six genes examined here. The other grasses are diversity averages from published studies (see review in ref. 1); however, they are often based on a limited number of loci.
Similar articles
- Post-domestication selection in the maize starch pathway.
Fan L, Bao J, Wang Y, Yao J, Gui Y, Hu W, Zhu J, Zeng M, Li Y, Xu Y. Fan L, et al. PLoS One. 2009 Oct 27;4(10):e7612. doi: 10.1371/journal.pone.0007612. PLoS One. 2009. PMID: 19859548 Free PMC article. - A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement.
Yamasaki M, Tenaillon MI, Bi IV, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD. Yamasaki M, et al. Plant Cell. 2005 Nov;17(11):2859-72. doi: 10.1105/tpc.105.037242. Epub 2005 Oct 14. Plant Cell. 2005. PMID: 16227451 Free PMC article. - Genetic diversity of maize kernel starch-synthesis genes with SNAPs.
Shin JH, Kwon SJ, Lee JK, Min HK, Kim NS. Shin JH, et al. Genome. 2006 Oct;49(10):1287-96. doi: 10.1139/g06-116. Genome. 2006. PMID: 17213911 - Genomic screening for artificial selection during domestication and improvement in maize.
Yamasaki M, Wright SI, McMullen MD. Yamasaki M, et al. Ann Bot. 2007 Nov;100(5):967-73. doi: 10.1093/aob/mcm173. Epub 2007 Aug 18. Ann Bot. 2007. PMID: 17704539 Free PMC article. Review. - Molecular and functional diversity of maize.
Buckler ES, Gaut BS, McMullen MD. Buckler ES, et al. Curr Opin Plant Biol. 2006 Apr;9(2):172-6. doi: 10.1016/j.pbi.2006.01.013. Epub 2006 Feb 3. Curr Opin Plant Biol. 2006. PMID: 16459128 Review.
Cited by
- Common gardens in teosintes reveal the establishment of a syndrome of adaptation to altitude.
Fustier MA, Martínez-Ainsworth NE, Aguirre-Liguori JA, Venon A, Corti H, Rousselet A, Dumas F, Dittberner H, Camarena MG, Grimanelli D, Ovaskainen O, Falque M, Moreau L, de Meaux J, Montes-Hernández S, Eguiarte LE, Vigouroux Y, Manicacci D, Tenaillon MI. Fustier MA, et al. PLoS Genet. 2019 Dec 20;15(12):e1008512. doi: 10.1371/journal.pgen.1008512. eCollection 2019 Dec. PLoS Genet. 2019. PMID: 31860672 Free PMC article. - Dissection of maize kernel composition and starch production by candidate gene association.
Wilson LM, Whitt SR, Ibáñez AM, Rocheford TR, Goodman MM, Buckler ES 4th. Wilson LM, et al. Plant Cell. 2004 Oct;16(10):2719-33. doi: 10.1105/tpc.104.025700. Epub 2004 Sep 17. Plant Cell. 2004. PMID: 15377761 Free PMC article. - The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model.
Wright SI, Charlesworth B. Wright SI, et al. Genetics. 2004 Oct;168(2):1071-6. doi: 10.1534/genetics.104.026500. Genetics. 2004. PMID: 15514076 Free PMC article. - A Maize CBM Domain Containing the Protein ZmCBM48-1 Positively Regulates Starch Synthesis in the Rice Endosperm.
Peng X, Yu W, Chen Y, Jiang Y, Ji Y, Chen L, Cheng B, Wu J. Peng X, et al. Int J Mol Sci. 2022 Jun 13;23(12):6598. doi: 10.3390/ijms23126598. Int J Mol Sci. 2022. PMID: 35743040 Free PMC article. - Genetic Diversity and Population Structure of Sorghum [Sorghum Bicolor (L.) Moench] Accessions as Revealed by Single Nucleotide Polymorphism Markers.
Enyew M, Feyissa T, Carlsson AS, Tesfaye K, Hammenhag C, Geleta M. Enyew M, et al. Front Plant Sci. 2022 Jan 5;12:799482. doi: 10.3389/fpls.2021.799482. eCollection 2021. Front Plant Sci. 2022. PMID: 35069657 Free PMC article.
References
- Buckler E S, IV, Thornsberry J M, Kresovich S. Genet Res. 2001;77:213–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources