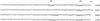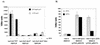Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter - PubMed (original) (raw)
Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter
Robin R Hulbert et al. J Bacteriol. 2002 Oct.
Abstract
The AraC homolog ToxT coordinately regulates virulence gene expression in Vibrio cholerae. ToxT is required for transcriptional activation of the genes encoding cholera toxin and the toxin coregulated pilus, among others. In this work we focused on the interaction of ToxT with the tcpA promoter and investigated the mechanism of ToxT-dependent transcriptional activation at tcpA. Deletion analysis showed that a region from -95 to +2 was sufficient for ToxT binding and activation, both of which were simultaneously lost when the deletion was extended to -63. A collection of point mutations generated by error-prone PCR revealed two small regions required for ToxT-dependent transactivation. Binding studies performed with representative mutations showed that the two regions define sites at which ToxT binds to the tcpA promoter region, most likely as a dimer. Results obtained by using a rpoA truncation mutation showed that ToxT-dependent activation at tcpA involves the C-terminal domain of the RNA polymerase alpha subunit. A model of ToxT-dependent transcriptional activation at tcpA is proposed, in which ToxT interacts with two A-rich regions of tcpA centered at -72 and -51 and requires the alpha C-terminal domain of RNA polymerase.
Figures
FIG. 1.
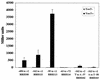
ToxT-dependent activation of tcpA-lacZ promoter deletions in E. coli. (A) Strains with no ToxT (shaded bars) or strains with the ToxT-expressing plasmid pTSS-5 (solid bars) were grown overnight in LB medium (pH 6.5) at 30°C. Promoter deletions are carried on plasmid pRS415. The values are averages for at least two independent experiments. (B) tcpA-lacZ deletion series. Most promoter deletions are within a promoter fragment that extends to position +2 relative to the start of transcription; the only exception is the −63 deletion, which extends to +72. The level of activation in the presence of ToxT is indicated for each deletion.
FIG. 2.
Sequences of single-point mutants with mutations in the region from −80 to −30, aligned with the sequence of the wild-type tcpA promoter. Base pair changes or deletions are shown; wild-type bases are indicated by dots. Strains carrying the tcpA-lacZ fusions on plasmid pRS415 with no ToxT or with the ToxT-expressing plasmid pTSS-5 were grown overnight in LB medium (pH 6.5) at 30°C. The β-galactosidase activity (in Miller units) and the level of activation (act.) in the presence of ToxT are shown for each strain. The values are averages for at least two independent experiments.
FIG. 3.
ToxT-dependent activation of representative tcpA-lacZ promoter constructs as λ lysogens integrated into the E. coli chromosome. Strains with no ToxT or strains with the ToxT-expressing plasmid pTSS-5 were grown overnight in LB medium (pH 6.5) at 30°C. The values are averages for at least two independent experiments.
FIG. 4.
Alignment of the tcpA promoter with other V. cholerae promoters regulated by ToxT. Sequences were aligned by using the −10 consensus sequence. The tcpI start site has been described previously (47), as have the tcpA start site (3) and the ctxA start site (51). The tagA start site is hypothetical.
FIG. 5.
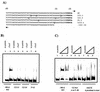
Six-His-tagged ToxT binds to tcpA promoter fragments in gel mobility shift assays. (A) Lanes 1 to 3, −95/+2 tcpA probe (lane 1, no protein; lane 2, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 3, 300 ng [9.3 pmol] of six-His-tagged ToxT); lanes 4 to 6, −63/+2 tcpA probe (lane 4, no protein; lane 5, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 6, 300 ng [9.3 pmol] of six-His-tagged ToxT); lanes 7 to 9, +1/+130 tcpP probe (lane 7, no protein; lane 8, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 9, 300 ng [9.3 pmol] of six-His-tagged ToxT). (B) Diagram showing the positions of tcpA promoter fragments used in the gel shift analysis. The −95/+2 probe includes both A tracts, while the −63/+2 probe includes one-half of the distal A tract and the entire proximal A tract.
FIG. 6.
Gel mobility shift assays with various tcpA promoter mutants. (A) Sequences of mutant probes used in binding studies. Base pair changes are underlined. Most of the probes are from −95 to +2; the only exception is the probe from −162 to −51. WT, wild type. (B) Lanes 1 and 2, −95/+2 tcpA probe (lane 1, no protein; lane 2, 150 ng [4.6 pmol] of six-His-tagged ToxT); lanes 3 and 4, CJ3.3 probe (lane 3, no protein; lane 4, 150 ng [4.6 pmol] of six-His-tagged ToxT); lanes 5 and 6, CJ2.6 probe (lane 5, no protein; lane 6, 150 ng [4.6 pmol] of six-His-tagged ToxT); lanes 7 and 8, F4.2 probe (lane 7, no protein; lane 8, 150 ng [4.6 pmol] of six-His-tagged ToxT). (C) Lanes 1 to 3, −95/+2 tcpA probe (lane 1, no protein; lane 2, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 3, 300 ng [9.3 pmol] of six-His-tagged ToxT); lanes 4 to 6, CJ10.4 probe (lane 4, no protein; lane 5, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 6, 300 ng [9.3 pmol] of six-His-tagged ToxT); lanes 7 to 9, site I probe (lane 7, no protein; lane 8, 150 ng [4.6 pmol] of six-His-tagged ToxT; lane 9, 300 ng [9.3 pmol] of six-His-tagged ToxT).
FIG. 7.
Effects of dominant negative RNA polymerase alpha subunit on tcpA-lacZ expression in V. cholerae (A) and E. coli (B). (A) Cultures of MBN135 (tcpA-lacZ), MBN142 (tcpA-lacZ Δ_toxT_), or MBN168 (tcpA-lacZ Δ_toxT_ Δ_hns_) carrying either pRH170 (full-length rpoA) or pRH171 (rpoA Δ235) were grown overnight in LB medium (pH 6.5) at 30°C with or without 0.04 mM IPTG. The values are averages for at least two independent experiments. WT, wild type. (B) Cultures of RRH115 (λ_tcpA-lacZ_ −162/+2) carrying no plasmid or pTSS-5 and either pRH170 or pRH171 were grown overnight in LB medium (pH 6.5) at 30°C with or without 0.04 mM IPTG. The values are averages for at least two independent experiments.
FIG. 8.
Model of transcriptional activation at tcpA. (A) During basal transcription, αCTD does not contact DNA. During activated transcription, a dimer of ToxT binds to site I (−82 to −64) and site II (−59 to −41). The ToxT molecule at site II recruits RNA polymerase through a mechanism involving the αCTD. See text for details. αNTD, alpha N-terminal domain. (B) Alignment of site I and site II at tcpA, tcpI, and tagA. Site I and site II at each promoter are underlined. The consensus sequence is shown at the bottom. An uppercase letter indicates that a base pair is present in all three promoters, and a lowercase letter indicates that base pair is conserved in two of the three promoters.
Comment in
- Growing repertoire of AraC/XylS activators.
Egan SM. Egan SM. J Bacteriol. 2002 Oct;184(20):5529-32. doi: 10.1128/JB.184.20.5529-5532.2002. J Bacteriol. 2002. PMID: 12270809 Free PMC article. Review. No abstract available.
Similar articles
- Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation.
Yu RR, DiRita VJ. Yu RR, et al. Mol Microbiol. 2002 Jan;43(1):119-34. doi: 10.1046/j.1365-2958.2002.02721.x. Mol Microbiol. 2002. PMID: 11849541 - The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT.
Withey JH, DiRita VJ. Withey JH, et al. Mol Microbiol. 2006 Mar;59(6):1779-89. doi: 10.1111/j.1365-2958.2006.05053.x. Mol Microbiol. 2006. PMID: 16553883 - pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae.
Behari J, Stagon L, Calderwood SB. Behari J, et al. J Bacteriol. 2001 Jan;183(1):178-88. doi: 10.1128/JB.183.1.178-188.2001. J Bacteriol. 2001. PMID: 11114915 Free PMC article. - The complexity of ToxT-dependent transcription in Vibrio cholerae.
Weber GG, Klose KE. Weber GG, et al. Indian J Med Res. 2011 Feb;133(2):201-6. Indian J Med Res. 2011. PMID: 21415495 Free PMC article. Review. - Regulation of virulence in Vibrio cholerae.
Klose KE. Klose KE. Int J Med Microbiol. 2001 May;291(2):81-8. doi: 10.1078/1438-4221-00104. Int J Med Microbiol. 2001. PMID: 11437342 Review.
Cited by
- Virstatin inhibits dimerization of the transcriptional activator ToxT.
Shakhnovich EA, Hung DT, Pierson E, Lee K, Mekalanos JJ. Shakhnovich EA, et al. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2372-7. doi: 10.1073/pnas.0611643104. Epub 2007 Feb 5. Proc Natl Acad Sci U S A. 2007. PMID: 17283330 Free PMC article. - Remodulation of bacterial transcriptome after acquisition of foreign DNA: the case of _irp_-HPI high-pathogenicity island in Vibrio anguillarum.
Lages MA, do Vale A, Lemos ML, Balado M. Lages MA, et al. mSphere. 2024 Jan 30;9(1):e0059623. doi: 10.1128/msphere.00596-23. Epub 2023 Dec 11. mSphere. 2024. PMID: 38078732 Free PMC article. - Identification and characterization of the functional toxboxes in the Vibrio cholerae cholera toxin promoter.
Dittmer JB, Withey JH. Dittmer JB, et al. J Bacteriol. 2012 Oct;194(19):5255-63. doi: 10.1128/JB.00952-12. Epub 2012 Jul 20. J Bacteriol. 2012. PMID: 22821976 Free PMC article. - Cyclic diguanylate regulates Vibrio cholerae virulence gene expression.
Tischler AD, Camilli A. Tischler AD, et al. Infect Immun. 2005 Sep;73(9):5873-82. doi: 10.1128/IAI.73.9.5873-5882.2005. Infect Immun. 2005. PMID: 16113306 Free PMC article. - Growing repertoire of AraC/XylS activators.
Egan SM. Egan SM. J Bacteriol. 2002 Oct;184(20):5529-32. doi: 10.1128/JB.184.20.5529-5532.2002. J Bacteriol. 2002. PMID: 12270809 Free PMC article. Review. No abstract available.
References
- Brown, R. C., and R. K. Taylor. 1995. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol. Microbiol. 16:425-439. - PubMed
- Cadwell, R. C., and G. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:26-33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases