FlgM gains structure in living cells - PubMed (original) (raw)
FlgM gains structure in living cells
Matthew M Dedmon et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Intrinsically disordered proteins such as FlgM play important roles in biology, but little is known about their structure in cells. We use NMR to show that FlgM gains structure inside living Escherichia coli cells and under physiologically relevant conditions in vitro, i.e., in solutions containing high concentrations (>/=400 g/liter) of glucose, BSA, or ovalbumin. Structure formation represents solute-induced changes in the equilibrium between the structured and disordered forms of FlgM. The results provide insight into how the environment of intrinsically disordered proteins could dictate their structure and, in turn, emphasize the relevance of studying proteins in living cells and in vitro under physiologically realistic conditions.
Figures
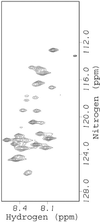
Figure 1
FlgM is structured in E. coli. The HSQC spectrum of FlgM in dilute solution (Left) and in living E. coli (Right). Red brackets surround some of the crosspeaks from the C-terminal half in FlgM that disappear in E. coli. The number near each bracket is the residue number (8). Dilute solution sample: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K. The E. coli spectrum was acquired as described by Serber et al. (9).
Figure 2
Glucose induces structure in FlgM. Overlaid HSQC spectra of FlgM in dilute solution (red) and in 450 g/liter 2.5 M glucose (black). Conditions: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K.
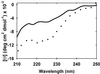
Figure 3
Glucose induces α-helix in FlgM. Far-UV CD spectra of 15 μM FlgM in dilute solution (solid line) and in 2.5 M (450 g/liter) glucose (dotted line). Both samples contain 10 mM sodium acetate (pH 5.0), 0.02% NaN3.
Figure 4
BSA induces structure in FlgM. HSQC spectrum of FlgM in 400 g/liter BSA. Conditions: 400 μM FlgM/10 mM sodium acetate, pH 5.0/10 mM NaCl/0.02% NaN3/10% (vol/vol) D2O at 298 K.
Similar articles
- Aquifex aeolicus FlgM protein exhibits a temperature-dependent disordered nature.
Molloy RG, Ma WK, Allen AC, Greenwood K, Bryan L, Sacora R, Williams L, Gage MJ. Molloy RG, et al. Biochim Biophys Acta. 2010 Jul;1804(7):1457-66. doi: 10.1016/j.bbapap.2010.03.002. Epub 2010 Mar 16. Biochim Biophys Acta. 2010. PMID: 20298817 - Intrinsically disordered protein from a pathogenic mesophile Mycobacterium tuberculosis adopts structured conformation at high temperature.
Kumar N, Shukla S, Kumar S, Suryawanshi A, Chaudhry U, Ramachandran S, Maiti S. Kumar N, et al. Proteins. 2008 May 15;71(3):1123-33. doi: 10.1002/prot.21798. Proteins. 2008. PMID: 18004752 - In-cell NMR of intrinsically disordered proteins in prokaryotic cells.
Ito Y, Mikawa T, Smith BO. Ito Y, et al. Methods Mol Biol. 2012;895:19-31. doi: 10.1007/978-1-61779-927-3_2. Methods Mol Biol. 2012. PMID: 22760309 - Bacterial in-cell NMR of human α-synuclein: a disordered monomer by nature?
Binolfi A, Theillet FX, Selenko P. Binolfi A, et al. Biochem Soc Trans. 2012 Oct;40(5):950-4. doi: 10.1042/BST20120096. Biochem Soc Trans. 2012. PMID: 22988846 Review. - The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations.
Daughdrill GW, Hanely LJ, Dahlquist FW. Daughdrill GW, et al. Biochemistry. 1998 Jan 27;37(4):1076-82. doi: 10.1021/bi971952t. Biochemistry. 1998. PMID: 9454599
Cited by
- Curcumin inhibits HIV-1 by promoting Tat protein degradation.
Ali A, Banerjea AC. Ali A, et al. Sci Rep. 2016 Jun 10;6:27539. doi: 10.1038/srep27539. Sci Rep. 2016. PMID: 27283735 Free PMC article. - Protecting activity of desiccated enzymes.
Piszkiewicz S, Gunn KH, Warmuth O, Propst A, Mehta A, Nguyen KH, Kuhlman E, Guseman AJ, Stadmiller SS, Boothby TC, Neher SB, Pielak GJ. Piszkiewicz S, et al. Protein Sci. 2019 May;28(5):941-951. doi: 10.1002/pro.3604. Epub 2019 Mar 30. Protein Sci. 2019. PMID: 30868674 Free PMC article. - Ultrafast Bioorthogonal Spin-Labeling and Distance Measurements in Mammalian Cells Using Small, Genetically Encoded Tetrazine Amino Acids.
Jana S, Evans EGB, Jang HS, Zhang S, Zhang H, Rajca A, Gordon SE, Zagotta WN, Stoll S, Mehl RA. Jana S, et al. J Am Chem Soc. 2023 Jul 12;145(27):14608-14620. doi: 10.1021/jacs.3c00967. Epub 2023 Jun 26. J Am Chem Soc. 2023. PMID: 37364003 Free PMC article. - Interplay of Structural Disorder and Short Binding Elements in the Cellular Chaperone Function of Plant Dehydrin ERD14.
Murvai N, Kalmar L, Szalaine Agoston B, Szabo B, Tantos A, Csikos G, Micsonai A, Kardos J, Vertommen D, Nguyen PN, Hristozova N, Lang A, Kovacs D, Buday L, Han KH, Perczel A, Tompa P. Murvai N, et al. Cells. 2020 Aug 7;9(8):1856. doi: 10.3390/cells9081856. Cells. 2020. PMID: 32784707 Free PMC article. - FlgM proteins from different bacteria exhibit different structural characteristics.
Ma WK, Hendrix R, Stewart C, Campbell EV, Lavarias M, Morris K, Nichol S, Gage MJ. Ma WK, et al. Biochim Biophys Acta. 2013 Apr;1834(4):808-16. doi: 10.1016/j.bbapap.2013.01.010. Epub 2013 Jan 22. Biochim Biophys Acta. 2013. PMID: 23352839 Free PMC article.
References
- Dunker A K, Brown C J, Lawson J D, Iakoucheva L M, Obradovic Z. Biochemistry. 2002;41:6573–6582. - PubMed
- Uversky V N, Gillespie J R, Fink A L. Proteins Struct Funct Genet. 2000;41:415–427. - PubMed
- Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Science. 1993;262:1277–1280. - PubMed
- Daughdrill G W, Hanely L J, Dahlquist F W. Biochemistry. 1998;37:1076–1082. - PubMed
- Schulman B A, Kim P S, Dobson C M, Redfield C. Nat Struct Biol. 1997;4:630–634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases