Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors - PubMed (original) (raw)
Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors
Jonathan R Pollack et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Genomic DNA copy number alterations are key genetic events in the development and progression of human cancers. Here we report a genome-wide microarray comparative genomic hybridization (array CGH) analysis of DNA copy number variation in a series of primary human breast tumors. We have profiled DNA copy number alteration across 6,691 mapped human genes, in 44 predominantly advanced, primary breast tumors and 10 breast cancer cell lines. While the overall patterns of DNA amplification and deletion corroborate previous cytogenetic studies, the high-resolution (gene-by-gene) mapping of amplicon boundaries and the quantitative analysis of amplicon shape provide significant improvement in the localization of candidate oncogenes. Parallel microarray measurements of mRNA levels reveal the remarkable degree to which variation in gene copy number contributes to variation in gene expression in tumor cells. Specifically, we find that 62% of highly amplified genes show moderately or highly elevated expression, that DNA copy number influences gene expression across a wide range of DNA copy number alterations (deletion, low-, mid- and high-level amplification), that on average, a 2-fold change in DNA copy number is associated with a corresponding 1.5-fold change in mRNA levels, and that overall, at least 12% of all the variation in gene expression among the breast tumors is directly attributable to underlying variation in gene copy number. These findings provide evidence that widespread DNA copy number alteration can lead directly to global deregulation of gene expression, which may contribute to the development or progression of cancer.
Figures
Figure 1
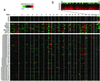
Genome-wide measurement of DNA copy number alteration by array CGH. (a) DNA copy number profiles are illustrated for cell lines containing different numbers of X chromosomes, for breast cancer cell lines, and for breast tumors. Each row represents a different cell line or tumor, and each column represents one of 6,691 different mapped human genes present on the microarray, ordered by genome map position from 1pter through Xqter. Moving average (symmetric 5-nearest neighbors) fluorescence ratios (test/reference) are depicted using a log2-based pseudocolor scale (indicated), such that red luminescence reflects fold-amplification, green luminescence reflects fold-deletion, and black indicates no change (gray indicates poorly measured data). (b) Enlarged view of DNA copy number profiles across the X chromosome, shown for cell lines containing different numbers of X chromosomes.
Figure 2
DNA copy number alteration across chromosome 8 by array CGH. (a) DNA copy number profiles are illustrated for cell lines containing different numbers of X chromosomes, for breast cancer cell lines, and for breast tumors. Breast cancer cell lines and tumors are separately ordered by hierarchical clustering to highlight recurrent copy number changes. The 241 genes present on the microarrays and mapping to chromosome 8 are ordered by position along the chromosome. Fluorescence ratios (test/reference) are depicted by a log2 pseudocolor scale (indicated). Selected genes are indicated with color-coded text (red, increased; green, decreased; black, no change; gray, not well measured) to reflect correspondingly altered mRNA levels (observed in the majority of the subset of samples displaying the DNA copy number change). The map positions for genes of interest that are not represented on the microarray are indicated in the row above those genes represented on the array. (b) Graphical display of DNA copy number profile for breast cancer cell line SKBR3. Fluorescence ratios (tumor/normal) are plotted on a log2 scale for chromosome 8 genes, ordered along the chromosome.
Figure 3
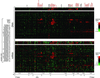
Concordance between DNA copy number and gene expression across chromosome 17. DNA copy number alteration (Upper) and mRNA levels (Lower) are illustrated for breast cancer cell lines and tumors. Breast cancer cell lines and tumors are separately ordered by hierarchical clustering (Upper), and the identical sample order is maintained (Lower). The 354 genes present on the microarrays and mapping to chromosome 17, and for which both DNA copy number and mRNA levels were determined, are ordered by position along the chromosome; selected genes are indicated in color-coded text (see Fig. 2 legend). Fluorescence ratios (test/reference) are depicted by separate log2 pseudocolor scales (indicated).
Figure 4
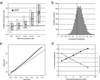
Genome-wide influence of DNA copy number alterations on mRNA levels. (a) For breast cancer cell lines (gray) and tumor samples (black), both mean-centered mRNA fluorescence ratio (log2 scale) quartiles (box plots indicate 25th, 50th, and 75th percentile) and averages (diamonds; _Y_-value error bars indicate standard errors of the mean) are plotted for each of five classes of genes, representing DNA deletion (tumor/normal ratio < 0.8), no change (0.8–1.2), low- (1.2–2), medium- (–4), and high-level (>4) amplification. P values for pair-wise Student's t tests, comparing averages between adjacent classes (moving left to right), are 4 × 10−49, 1 × 10−49, 5 × 10−5, 1 × 10−2 (cell lines), and 1 × 10−43, 1 × 10−214, 5 × 10−41, 1 × 10−4 (tumors). (b) Distribution of correlations between DNA copy number and mRNA levels, for 6,095 different human genes across 37 breast tumor samples. (c) Plot of observed versus expected correlation coefficients. The expected values were obtained by randomization of the sample labels in the DNA copy number data set. The line of unity is indicated. (d) Percent variance in gene expression (among tumors) directly explained by variation in gene copy number. Percent variance explained (black line) and fraction of data retained (gray line) are plotted for different fluorescence intensity/background (a rough surrogate for signal/noise) cutoff values. Fraction of data retained is relative to the 1.2 intensity/background cutoff. Details of the linear regression model used to estimate the fraction of variation in gene expression attributable to underlying DNA copy number alteration can be found in the supporting information (see Estimating the Fraction of Variation in Gene Expression Attributable to Underlying DNA Copy Number Alteration).
Similar articles
- Impact of DNA amplification on gene expression patterns in breast cancer.
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A. Hyman E, et al. Cancer Res. 2002 Nov 1;62(21):6240-5. Cancer Res. 2002. PMID: 12414653 - High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays.
Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. Pinkel D, et al. Nat Genet. 1998 Oct;20(2):207-11. doi: 10.1038/2524. Nat Genet. 1998. PMID: 9771718 - New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays.
Kauraniemi P, Bärlund M, Monni O, Kallioniemi A. Kauraniemi P, et al. Cancer Res. 2001 Nov 15;61(22):8235-40. Cancer Res. 2001. PMID: 11719455 - The 17q23 amplicon and breast cancer.
Sinclair CS, Rowley M, Naderi A, Couch FJ. Sinclair CS, et al. Breast Cancer Res Treat. 2003 Apr;78(3):313-22. doi: 10.1023/a:1023081624133. Breast Cancer Res Treat. 2003. PMID: 12755490 Review. - Profiling breast cancer by array CGH.
Albertson DG. Albertson DG. Breast Cancer Res Treat. 2003 Apr;78(3):289-98. doi: 10.1023/a:1023025506386. Breast Cancer Res Treat. 2003. PMID: 12755488 Review.
Cited by
- Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma.
Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Nishimura Y, et al. Br J Cancer. 2013 Apr 2;108(6):1324-31. doi: 10.1038/bjc.2013.65. Epub 2013 Feb 19. Br J Cancer. 2013. PMID: 23422756 Free PMC article. - CDX2 is an amplified lineage-survival oncogene in colorectal cancer.
Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S, Ko ME, Lin AY, van de Rijn M, Pollack JR. Salari K, et al. Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):E3196-205. doi: 10.1073/pnas.1206004109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112155 Free PMC article. - Laminin-binding integrin gene copy number alterations in distinct epithelial-type cancers.
Harryman WL, Pond E, Singh P, Little AS, Eschbacher JM, Nagle RB, Cress AE. Harryman WL, et al. Am J Transl Res. 2016 Feb 15;8(2):940-54. eCollection 2016. Am J Transl Res. 2016. PMID: 27158381 Free PMC article. - Amplification and high-level expression of heat shock protein 90 marks aggressive phenotypes of human epidermal growth factor receptor 2 negative breast cancer.
Cheng Q, Chang JT, Geradts J, Neckers LM, Haystead T, Spector NL, Lyerly HK. Cheng Q, et al. Breast Cancer Res. 2012 Apr 17;14(2):R62. doi: 10.1186/bcr3168. Breast Cancer Res. 2012. PMID: 22510516 Free PMC article. - Inference of tumor phylogenies from genomic assays on heterogeneous samples.
Subramanian A, Shackney S, Schwartz R. Subramanian A, et al. J Biomed Biotechnol. 2012;2012:797812. doi: 10.1155/2012/797812. Epub 2012 May 13. J Biomed Biotechnol. 2012. PMID: 22654484 Free PMC article.
References
- Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. - PubMed
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O P. Genes Chromosomes Cancer. 1998;21:177–184. - PubMed
- Forozan F, Mahlamaki E H, Monni O, Chen Y, Veldman R, Jiang Y, Gooden G C, Ethier S P, Kallioniemi A, Kallioniemi O P. Cancer Res. 2000;60:4519–4525. - PubMed
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Genes Chromosomes Cancer. 1997;20:399–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases