Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment - PubMed (original) (raw)
Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment
Alexander Loy et al. Appl Environ Microbiol. 2002 Oct.
Abstract
For cultivation-independent detection of sulfate-reducing prokaryotes (SRPs) an oligonucleotide microarray consisting of 132 16S rRNA gene-targeted oligonucleotide probes (18-mers) having hierarchical and parallel (identical) specificity for the detection of all known lineages of sulfate-reducing prokaryotes (SRP-PhyloChip) was designed and subsequently evaluated with 41 suitable pure cultures of SRPs. The applicability of SRP-PhyloChip for diversity screening of SRPs in environmental and clinical samples was tested by using samples from periodontal tooth pockets and from the chemocline of a hypersaline cyanobacterial mat from Solar Lake (Sinai, Egypt). Consistent with previous studies, SRP-PhyloChip indicated the occurrence of Desulfomicrobium spp. in the tooth pockets and the presence of Desulfonema- and Desulfomonile-like SRPs (together with other SRPs) in the chemocline of the mat. The SRP-PhyloChip results were confirmed by several DNA microarray-independent techniques, including specific PCR amplification, cloning, and sequencing of SRP 16S rRNA genes and the genes encoding the dissimilatory (bi)sulfite reductase (dsrAB).
Figures
FIG. 1.
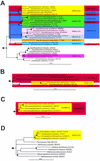
Phylogenetic affiliations of SRPs belonging to the orders “Desulfobacterales_” and “_Syntrophobacterales_” of the class “_Deltaproteobacteria.” The 16S rRNA consensus tree was constructed from comparative sequence analysis data by using maximum-parsimony, maximum-likelihood, and neighbor-joining methods and applying filters excluding all alignment positions which are not conserved in at least 50% of all bacterial and deltaproteobacterial 16S rRNA sequences. A collection of organisms representing all major lineages of the Archaea and Bacteria was used as an outgroup. Multifurcations connect branches for which a relative order could not be determined unambiguously. Non-SRPs are underlined. Parsimony bootstrap values (1,000 resamplings) for branches are indicated by solid circles (>90%) or an open circle (75 to 90%). Branches without circles had bootstrap values of less than 75%. The bar indicates 10% estimated sequence divergence (distance inferred by neighbor joining by using a 50% bacterial conservation filter). The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. Probes SRB385Db, DSS658, DSR651, and DSB804 are not shown to enhance clarity.
FIG. 2.
Phylogenetic affiliations of SRPs belonging to the order “Desulfovibrionales_” of the class “_Deltaproteobacteria.” The 16S rRNA consensus tree was constructed as described in the legend to Fig. 1. Non-SRPs are underlined. The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. Probes SRB385, DSV1292, and DSV698 are not shown to enhance clarity.
FIG. 3.
(A) Phylogenetic affiliations of SRPs belonging to the family Peptococcaceae of the phylum Firmicutes (low-G+C-content gram-positive bacteria). (B) Phylogenetic affiliations of SRPs belonging to the genus Thermodesulfovibrio of the phylum Nitrospira. (C) Phylogenetic affiliations of SRPs belonging to the phylum Thermodesulfobacteria. (D) Phylogenetic affiliations of SRPs of the genus Archaeoglobus belonging to the phylum Euryarchaeota. In all panels non-SRPs are underlined. The 16S rRNA consensus trees were constructed as described in the legend to Fig. 1. The colored boxes show the specificities (perfect-match target organisms) of the SRP-PhyloChip probes (indicated by short names). The numbers of probes with identical specificities for the target organisms are indicated in parentheses. In panel A probes DFMI210 and DFMI229 are not shown to enhance clarity.
FIG. 4.
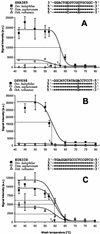
Melting curves for probe SRB385 (A), probe DSV698 (B), and probe EUB338 (C) after hybridization with fluorescently labeled PCR-amplified 16S rRNA gene fragments of Desulfovibrio halophilus, Desulfomicrobium aspheronum, and Desulfohalobium retbaense. For each probe the difference alignment with these reference SRPs is shown. The observed dissociation temperature (Td) is indicated for each probe. Each data point represents the mean signal intensity value for 10 probe spots (local background was subtracted for each measurement). The error bars indicate the standard deviations. For each wash temperature and reference organism a separate microarray hybridization was performed. a.u., arbitrary units.
FIG. 5.
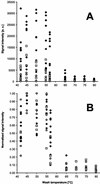
Hybridization intensities of probes forming perfect-match (diamonds), one-mismatch (squares), and two-mismatch (circles) duplexes after hybridization with fluorescently labeled PCR-amplified 16S rRNA gene fragments of Desulfovibrio halophilus at different stringencies. (A) Mean signal intensities (for 10 spots, with local background subtracted) for each probe and wash temperature. (B) Normalized mean signal intensity values for each probe and wash temperature. Mean intensity values were normalized for each probe separately by assuming that the highest value observed at the different wash temperatures had a value of 1.00. In panel B, probes which showed no hybridization signals at low stringencies are not shown.
FIG. 6.
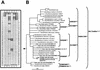
(A) Use of the SRP-PhyloChip for surveys of SRP diversity in periodontal tooth pockets. On the microarray each probe was spotted in duplicate. For each microarray position, the probe sequence and specificity are shown in Table 3. Probe spots having a signal-to-noise ratio equal to or greater than 2.0 are indicated by boldface boxes and were considered to be positive. (B) Evaluation of the microarray results by amplification, cloning, and comparative sequence analysis of 16S rRNA gene fragments by using _Desulfomicrobium_-specific primers for PCR. 16S rRNA gene clones obtained from the tooth pockets are indicated by boldface type. The tree is based on a maximum-likelihood analysis performed with a 50% conservation filter for the Bacteria. Multifurcations connect branches for which a relative order could not be determined unambiguously after different treeing methods and filters were used. The bar indicates 10% estimated sequence divergence. The brackets indicate the perfect-match target organisms for the probes. The microarray position is indicated after each probe name.
FIG. 7.
(A) Use of the SRP-PhyloChip for surveys of SRP diversity in the chemocline of a cyanobacterial microbial mat. On the microarray each probe was spotted in duplicate. For each microarray position, the probe sequence and specificity are shown in Table 3. Probe spots having a signal-to-noise ratio equal to or greater than 2.0 are indicated by boldface boxes and were considered to be positive. The dotted boldface boxes indicate that only one of the duplicate spots had a signal-to-noise ratio equal to or greater than 2.0. (B) Evaluation of the microarray results by amplification, cloning, and comparative sequence analysis of 16S rRNA gene fragments by using primers specific for some Desulfonema species (SLM-DSN clones) and most members of the “_Desulfobacterales_” and “_Syntrophobacterales_” (SLM-DSBAC clones). Clone SLM-CP-116 was obtained from the mat chemocline by amplification, cloning, and sequencing after enrichment by using probe DSN658 as the capture probe. 16S rRNA gene clones obtained from the chemocline of the Solar Lake mat are indicated by boldface type. The tree is based on a maximum-likelihood analysis performed with a 50% conservation filter for the Bacteria. Multifurcations connect branches for which a relative order could not be determined unambiguously after different treeing methods and filters were used. The bar indicates 10% estimated sequence divergence. The brackets indicate the perfect-match target organisms of the probes. The microarray position is indicated after each probe name. The amplified and sequenced 16S rRNA gene fragment of Solar Lake mat clone SLM-DSBAC-74 (indicated by an asterisk) is outside the target site for probe DSMON95 and has one mismatch (located at position 16) within the target site for probe DSMON1421.
References
- Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous