Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? - PubMed (original) (raw)
. 2002 Oct 15;99(21):13442-7.
doi: 10.1073/pnas.212410399. Epub 2002 Oct 1.
Sandrine Poncet, Anne Galinier, Vicente Monedero, Sonia Fieulaine, Joël Janin, Sylvie Nessler, José Antonio Marquez, Klaus Scheffzek, Sonja Hasenbein, Wolfgang Hengstenberg, Josef Deutscher
Affiliations
- PMID: 12359880
- PMCID: PMC129692
- DOI: 10.1073/pnas.212410399
Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life?
Ivan Mijakovic et al. Proc Natl Acad Sci U S A. 2002.
Abstract
In most Gram-positive bacteria, serine-46-phosphorylated HPr (P-Ser-HPr) controls the expression of numerous catabolic genes ( approximately 10% of their genome) by acting as catabolite corepressor. HPr kinase/phosphorylase (HprK/P), the bifunctional sensor enzyme for catabolite repression, phosphorylates HPr, a phosphocarrier protein of the sugar-transporting phosphoenolpyruvate/glycose phosphotransferase system, in the presence of ATP and fructose-1,6-bisphosphate but dephosphorylates P-Ser-HPr when phosphate prevails over ATP and fructose-1,6-bisphosphate. We demonstrate here that P-Ser-HPr dephosphorylation leads to the formation of HPr and pyrophosphate. HprK/P, which binds phosphate at the same site as the beta phosphate of ATP, probably uses the inorganic phosphate to carry out a nucleophilic attack on the phosphoryl bond in P-Ser-HPr. HprK/P is the first enzyme known to catalyze P-protein dephosphorylation via this phospho-phosphorolysis mechanism. This reaction is reversible, and at elevated pyrophosphate concentrations, HprK/P can use pyrophosphate to phosphorylate HPr. Growth of Bacillus subtilis on glucose increased intracellular pyrophosphate to concentrations ( approximately 6 mM), which in in vitro tests allowed efficient pyrophosphate-dependent HPr phosphorylation. To effectively dephosphorylate P-Ser-HPr when glucose is exhausted, the pyrophosphate concentration in the cells is lowered to 1 mM. In B. subtilis, this might be achieved by YvoE. This protein exhibits pyrophosphatase activity, and its gene is organized in an operon with hprK.
Figures
Figure 1
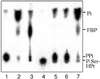
[32P]P-Ser-HPr dephosphorylation by B. subtilis HprK/P. [32P]P-Ser-HPr dephosphorylation was carried out as described in Materials and Methods. The reaction products were separated by TLC with 0.3 M potassium phosphate, pH 7.4. Radioactive standards for pyrophosphate, phosphate, and P-Ser-HPr (lanes 1–3, respectively); 10 μM [32P]P-Ser-HPr incubated with HprK/P for 30, 2, 5, 10, 20, and 30 min (lanes 4–9, respectively) in the absence (lane 4) or in the presence (lanes 5–9) of 5 mM phosphate; lane 10, 0.1 mM [32P]P-Ser-HPr was incubated for 15 min with HprK/P in the presence of 20 μM phosphate, and lane 11, same experiment as in lane 10 except that before adding HprK/P the sample was preincubated for 10 min with maltose and maltose phosphorylase. PPi, pyrophosphate.
Figure 2
Identification of the [32P]P-Ser-HPr dephosphorylation product. The reaction products of HprK/P-catalyzed [32P]P-Ser-HPr dephosphorylation were separated by TLC using 0.3 M KH2PO4 as solvent. Radioactive standards for pyrophosphate and P-Ser-HPr (lanes 1 and 4, respectively); 1-[32P]FBP formed from fructose-6-P and either [32P]pyrophosphate or [γ-32P]ATP (lanes 2 and 3, respectively). Dephosphorylation of [32P]P-Ser-HPr with B. subtilis HprK/P was carried out in the presence of fructose-6-P (lane 5), pyrophosphate-dependent phosphofructokinase (lane 6), and fructose-6-P and pyrophosphate-dependent phosphofructokinase (lane 7).
Figure 3
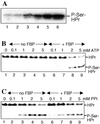
Pyrophosphate-dependent HprK/P-catalyzed phosphorylation of HPr. (A) HPr was phosphorylated with B. subtilis HprK/P and [32P]pyrophosphate at various concentrations (0.1, 0.2, 0.4, 1, 2 ,and 4 mM for lanes 1–6, respectively). The samples were separated on a 12.5% polyacrylamide gel containing 0.1% SDS. (B and C) HPr was phosphorylated with B. subtilis HprK/P in the absence (lanes 1–5) or presence (lanes 6–9) of 5 mM FBP with varying concentrations of ATP (B) or pyrophosphate (C), and samples were separated on nondenaturing polyacrylamide gels.
Figure 4
YvoE-catalyzed hydrolysis of pyrophosphate. [32P]Pyrophosphate (10 μM) was incubated with 0, 2.5, 25, and 50 nM YvoE (lanes 1–4, respectively). Phosphorolysis of 0.1 mM [32P]P-Ser-HP by HprK/P in the absence of YvoE (lane 5) and in the presence of 250 nM YvoE (lane 6). Samples were separated by TLC.
Similar articles
- X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Fieulaine S, Morera S, Poncet S, Mijakovic I, Galinier A, Janin J, Deutscher J, Nessler S. Fieulaine S, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13437-41. doi: 10.1073/pnas.192368699. Epub 2002 Oct 1. Proc Natl Acad Sci U S A. 2002. PMID: 12359875 Free PMC article. - Phosphorylation of HPr by the bifunctional HPr Kinase/P-ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion.
Dossonnet V, Monedero V, Zagorec M, Galinier A, Pérez-Martínez G, Deutscher J. Dossonnet V, et al. J Bacteriol. 2000 May;182(9):2582-90. doi: 10.1128/JB.182.9.2582-2590.2000. J Bacteriol. 2000. PMID: 10762262 Free PMC article. - HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria.
Poncet S, Mijakovic I, Nessler S, Gueguen-Chaignon V, Chaptal V, Galinier A, Boël G, Mazé A, Deutscher J. Poncet S, et al. Biochim Biophys Acta. 2004 Mar 11;1697(1-2):123-35. doi: 10.1016/j.bbapap.2003.11.018. Biochim Biophys Acta. 2004. PMID: 15023355 Review. - Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC.
Singh KD, Halbedel S, Görke B, Stülke J. Singh KD, et al. J Mol Microbiol Biotechnol. 2007;13(1-3):165-71. doi: 10.1159/000103608. J Mol Microbiol Biotechnol. 2007. PMID: 17693724 - Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria.
Boël G, Mijakovic I, Mazé A, Poncet S, Taha MK, Larribe M, Darbon E, Khemiri A, Galinier A, Deutscher J. Boël G, et al. J Mol Microbiol Biotechnol. 2003;5(4):206-15. doi: 10.1159/000071072. J Mol Microbiol Biotechnol. 2003. PMID: 12867744 Review.
Cited by
- HPr kinase/phosphorylase, the sensor enzyme of catabolite repression in Gram-positive bacteria: structural aspects of the enzyme and the complex with its protein substrate.
Nessler S, Fieulaine S, Poncet S, Galinier A, Deutscher J, Janin J. Nessler S, et al. J Bacteriol. 2003 Jul;185(14):4003-10. doi: 10.1128/JB.185.14.4003-4010.2003. J Bacteriol. 2003. PMID: 12837773 Free PMC article. No abstract available. - A Review of the Bacterial Phosphoproteomes of Beneficial Microbes.
Lim S. Lim S. Microorganisms. 2023 Apr 3;11(4):931. doi: 10.3390/microorganisms11040931. Microorganisms. 2023. PMID: 37110354 Free PMC article. Review. - Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis.
Musumeci L, Bongiorni C, Tautz L, Edwards RA, Osterman A, Perego M, Mustelin T, Bottini N. Musumeci L, et al. J Bacteriol. 2005 Jul;187(14):4945-56. doi: 10.1128/JB.187.14.4945-4956.2005. J Bacteriol. 2005. PMID: 15995210 Free PMC article. - Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources.
Singh KD, Schmalisch MH, Stülke J, Görke B. Singh KD, et al. J Bacteriol. 2008 Nov;190(21):7275-84. doi: 10.1128/JB.00848-08. Epub 2008 Aug 29. J Bacteriol. 2008. PMID: 18757537 Free PMC article. - High-resolution structure of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus aureus and characterization of its interaction with the bifunctional HPr kinase/phosphorylase.
Maurer T, Meier S, Kachel N, Munte CE, Hasenbein S, Koch B, Hengstenberg W, Kalbitzer HR. Maurer T, et al. J Bacteriol. 2004 Sep;186(17):5906-18. doi: 10.1128/JB.186.17.5906-5918.2004. J Bacteriol. 2004. PMID: 15317796 Free PMC article.
References
- Mason P W, Carbone D P, Cushman R A, Waggoner A S. J Biol Chem. 1981;256:1861–1866. - PubMed
- Neves A R, Ramos A, Nunes M C, Kleerebezem M, Hugenholtz J, de Vos W M, Almeida J, Santos H. Biotechnol Bioeng. 1998;64:200–212. - PubMed
- Deutscher J, Galinier A, Martin-Verstraete I. In: Bacillus subtilis and its Closest Relatives: From Genes to Cells. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 2001. pp. 129–150.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases