SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines - PubMed (original) (raw)
Comparative Study
SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines
Ada Ching et al. BMC Genet. 2002.
Abstract
Background: Recent studies of ancestral maize populations indicate that linkage disequilibrium tends to dissipate rapidly, sometimes within 100 bp. We set out to examine the linkage disequilibrium and diversity in maize elite inbred lines, which have been subject to population bottlenecks and intense selection by breeders. Such population events are expected to increase the amount of linkage disequilibrium, but reduce diversity. The results of this study will inform the design of genetic association studies.
Results: We examined the frequency and distribution of DNA polymorphisms at 18 maize genes in 36 maize inbreds, chosen to represent most of the genetic diversity in U.S. elite maize breeding pool. The frequency of nucleotide changes is high, on average one polymorphism per 31 bp in non-coding regions and 1 polymorphism per 124 bp in coding regions. Insertions and deletions are frequent in non-coding regions (1 per 85 bp), but rare in coding regions. A small number (2-8) of distinct and highly diverse haplotypes can be distinguished at all loci examined. Within genes, SNP loci comprising the haplotypes are in linkage disequilibrium with each other.
Conclusions: No decline of linkage disequilibrium within a few hundred base pairs was found in the elite maize germplasm. This finding, as well as the small number of haplotypes, relative to neutral expectation, is consistent with the effects of breeding-induced bottlenecks and selection on the elite germplasm pool. The genetic distance between haplotypes is large, indicative of an ancient gene pool and of possible interspecific hybridization events in maize ancestry.
Figures
Figure 1
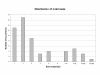
Distribution of insertion /deletion sizes Number of observed insertion / deletion polymorphisms (indels) of each size class is shown.
Figure 2
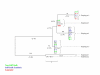
Neighbor-joining trees representing Adh1 haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
Figure 3
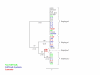
Neighbor-joining trees representing stearoyl-ACP desaturase haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
Figure 4
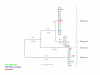
Neighbor-joining trees representing acetyl-CoA C-acyltransferase haplotype relationships. Level of support for branch points is indicated in %, and branch length expressed as nucleotide differences are shown in parentheses. Genotypes correspond to those of Table 1, and color indicates major heterotic groups: stiff stalk (blue), non stiff stalk (green) and Lancaster (red).
Figure 5
Composite plot of linkage disequilibrium as a function of distance. Two measures of linkage disequilibrium, absolute value of D' (A) and r2 (B) are shown as a function of distance for all loci examined. LD values between all pairs of SNP were plotted. Logarithmic trend line is included in plot (B). Of the 344 pairwise comparisons, 161 were significant at P < 0.01, with Bonferroni correction, and 126 were significant at P < 0.001 level.
Similar articles
- Single nucleotide polymorphisms and linkage disequilibrium in sunflower.
Kolkman JM, Berry ST, Leon AJ, Slabaugh MB, Tang S, Gao W, Shintani DK, Burke JM, Knapp SJ. Kolkman JM, et al. Genetics. 2007 Sep;177(1):457-68. doi: 10.1534/genetics.107.074054. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660563 Free PMC article. - Corn and humans: recombination and linkage disequilibrium in two genomes of similar size.
Rafalski A, Morgante M. Rafalski A, et al. Trends Genet. 2004 Feb;20(2):103-11. doi: 10.1016/j.tig.2003.12.002. Trends Genet. 2004. PMID: 14746992 Review. - Linkage disequilibrium and sequence diversity in a 500-kbp region around the adh1 locus in elite maize germplasm.
Jung M, Ching A, Bhattramakki D, Dolan M, Tingey S, Morgante M, Rafalski A. Jung M, et al. Theor Appl Genet. 2004 Aug;109(4):681-9. doi: 10.1007/s00122-004-1695-8. Theor Appl Genet. 2004. PMID: 15300382 - Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers.
Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J. Yan J, et al. PLoS One. 2009 Dec 24;4(12):e8451. doi: 10.1371/journal.pone.0008451. PLoS One. 2009. PMID: 20041112 Free PMC article. - On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.
Garner C, Slatkin M. Garner C, et al. Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
- Experimental evaluation of effectiveness of genomic selection for resistance to northern corn leaf blight in maize.
Lohithaswa HC, Balasundara DC, Mallikarjuna MG, Sowmya MS, Mallikarjuna N, Kulkarni RS, Pandravada AS, Bhatia BS. Lohithaswa HC, et al. J Appl Genet. 2024 Oct 24. doi: 10.1007/s13353-024-00911-x. Online ahead of print. J Appl Genet. 2024. PMID: 39446310 - Genome-wide association study reveals SNP markers controlling drought tolerance and related agronomic traits in chickpea across multiple environments.
Istanbuli T, Nassar AE, Abd El-Maksoud MM, Tawkaz S, Alsamman AM, Hamwieh A. Istanbuli T, et al. Front Plant Sci. 2024 Mar 8;15:1260690. doi: 10.3389/fpls.2024.1260690. eCollection 2024. Front Plant Sci. 2024. PMID: 38525151 Free PMC article. - ZmARF1 positively regulates low phosphorus stress tolerance via modulating lateral root development in maize.
Wu F, Yahaya BS, Gong Y, He B, Gou J, He Y, Li J, Kang Y, Xu J, Wang Q, Feng X, Tang Q, Liu Y, Lu Y. Wu F, et al. PLoS Genet. 2024 Feb 5;20(2):e1011135. doi: 10.1371/journal.pgen.1011135. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38315718 Free PMC article. - Genome-wide association study of maize resistance to Pythium aristosporum stalk rot.
Hou M, Cao Y, Zhang X, Zhang S, Jia T, Yang J, Han S, Wang L, Li J, Wang H, Zhang L, Wu X, Duan C, Li H. Hou M, et al. Front Plant Sci. 2023 Aug 17;14:1239635. doi: 10.3389/fpls.2023.1239635. eCollection 2023. Front Plant Sci. 2023. PMID: 37662167 Free PMC article. - Identification of Genomic Regions Associated with Agronomic and Disease Resistance Traits in a Large Set of Multiple DH Populations.
Sadessa K, Beyene Y, Ifie BE, Suresh LM, Olsen MS, Ogugo V, Wegary D, Tongoona P, Danquah E, Offei SK, Prasanna BM, Gowda M. Sadessa K, et al. Genes (Basel). 2022 Feb 15;13(2):351. doi: 10.3390/genes13020351. Genes (Basel). 2022. PMID: 35205395 Free PMC article.
References
- Lindblad-Toh K, Winchester E, Daly M, Wang D, Hirschhorn JN, Laviolette JP, Ardlie K, Reich DE, Robinson E, Sklar P, Shah N, Thomas D, Fan JB, Gingeras T, Warrington J, Patil N, Hudson TJ, Lander ES. Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat Genet. 2000;24:381–386. doi: 10.1038/74215. - DOI - PubMed
- Bhattramakki D, Rafalski A. Discovery and Application of Single Nucleotide Polymorphism Markers in Plants. In: Henry RJ, editor. Plant Genotyping: The DNA Fingerprinting of Plants. Wallingford Oxon, UK: CABI Publishing; 2001.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources