The generic genome browser: a building block for a model organism system database - PubMed (original) (raw)
The generic genome browser: a building block for a model organism system database
Lincoln D Stein et al. Genome Res. 2002 Oct.
Abstract
The Generic Model Organism System Database Project (GMOD) seeks to develop reusable software components for model organism system databases. In this paper we describe the Generic Genome Browser (GBrowse), a Web-based application for displaying genomic annotations and other features. For the end user, features of the browser include the ability to scroll and zoom through arbitrary regions of a genome, to enter a region of the genome by searching for a landmark or performing a full text search of all features, and the ability to enable and disable tracks and change their relative order and appearance. The user can upload private annotations to view them in the context of the public ones, and publish those annotations to the community. For the data provider, features of the browser software include reliance on readily available open source components, simple installation, flexible configuration, and easy integration with other components of a model organism system Web site. GBrowse is freely available under an open source license. The software, its documentation, and support are available at http://www.gmod.org.
Figures
Figure 1
The user enters GBrowse by typing a landmark name into the text field at top. Landmarks can be gene names, clone names, accession numbers, or any other identifier configured by the administrator. Once a region is selected, it is displayed in a detailed view that summarizes annotations and other genomic features. An overview panel and a navigation bar together allow the user to move from one place to another.
Figure 2
The detailed view after zooming out to 200 kb, showing semantic zooming.
Figure 3
A search for the term “7 transmembrane receptor.”
Figure 4
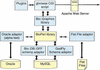
The Generic Genome Browser is built from multiple software modules. In this illustration, modules that were not produced as part of this project are shown in a lighter color.
Figure 5
The Bio::DB::GFF database uses a minimal schema to represent features on sequences. The main tables are fdata, which contains the position and type of each feature, fgroup, which tracks the grouping of subfeatures into features, such as high-similarity pairs in a gapped alignment, fdna, which stores the raw DNA sequence, and fattribute_to_feature, which allows attribute information to be attached to features. Attributes are used for storing such textual information as notes, synonyms, and evidence codes. The fattribute and ftype tables, respectively, hold attribute names and the method and source fields. For retrieval efficiency, the fdna table fragments each DNA into small pieces and stores the beginning of each piece in the foffset field.
Figure 6
Creating a new track of targeted deletions using GBrowse.
Similar articles
- Using the Generic Synteny Browser (GBrowse_syn).
McKay SJ, Vergara IA, Stajich JE. McKay SJ, et al. Curr Protoc Bioinformatics. 2010 Sep;Chapter 9:Unit 9.12. doi: 10.1002/0471250953.bi0912s31. Curr Protoc Bioinformatics. 2010. PMID: 20836076 Free PMC article. - Using the Generic Genome Browser (GBrowse).
Donlin MJ. Donlin MJ. Curr Protoc Bioinformatics. 2009 Dec;Chapter 9:Unit 9.9. doi: 10.1002/0471250953.bi0909s28. Curr Protoc Bioinformatics. 2009. PMID: 19957275 - SynBrowse: a synteny browser for comparative sequence analysis.
Pan X, Stein L, Brendel V. Pan X, et al. Bioinformatics. 2005 Sep 1;21(17):3461-8. doi: 10.1093/bioinformatics/bti555. Epub 2005 Jun 30. Bioinformatics. 2005. PMID: 15994196 - Using the Generic Genome Browser (GBrowse).
Donlin MJ. Donlin MJ. Curr Protoc Bioinformatics. 2007 Mar;Chapter 9:Unit 9.9. doi: 10.1002/0471250953.bi0909s17. Curr Protoc Bioinformatics. 2007. PMID: 18428797 - FlyBase : a database for the Drosophila research community.
Drysdale R; FlyBase Consortium. Drysdale R, et al. Methods Mol Biol. 2008;420:45-59. doi: 10.1007/978-1-59745-583-1_3. Methods Mol Biol. 2008. PMID: 18641940 Review.
Cited by
- The YeastGenome app: the Saccharomyces Genome Database at your fingertips.
Wong ED, Karra K, Hitz BC, Hong EL, Cherry JM. Wong ED, et al. Database (Oxford). 2013 Feb 8;2013:bat004. doi: 10.1093/database/bat004. Print 2013. Database (Oxford). 2013. PMID: 23396302 Free PMC article. - The Eimeria transcript DB: an integrated resource for annotated transcripts of protozoan parasites of the genus Eimeria.
Rangel LT, Novaes J, Durham AM, Madeira AM, Gruber A. Rangel LT, et al. Database (Oxford). 2013 Feb 14;2013:bat006. doi: 10.1093/database/bat006. Print 2013. Database (Oxford). 2013. PMID: 23411718 Free PMC article. - BESC knowledgebase public portal.
Syed MH, Karpinets TV, Parang M, Leuze MR, Park BH, Hyatt D, Brown SD, Moulton S, Galloway MD, Uberbacher EC. Syed MH, et al. Bioinformatics. 2012 Mar 1;28(5):750-1. doi: 10.1093/bioinformatics/bts016. Epub 2012 Jan 11. Bioinformatics. 2012. PMID: 22238270 Free PMC article. - Defining bacterial regulons using ChIP-seq.
Myers KS, Park DM, Beauchene NA, Kiley PJ. Myers KS, et al. Methods. 2015 Sep 15;86:80-8. doi: 10.1016/j.ymeth.2015.05.022. Epub 2015 May 29. Methods. 2015. PMID: 26032817 Free PMC article. Review. - Translational arrest due to cytoplasmic redox stress delays adaptation to growth on methanol and heterologous protein expression in a typical fed-batch culture of Pichia pastoris.
Edwards-Jones B, Aw R, Barton GR, Tredwell GD, Bundy JG, Leak DJ. Edwards-Jones B, et al. PLoS One. 2015 Mar 18;10(3):e0119637. doi: 10.1371/journal.pone.0119637. eCollection 2015. PLoS One. 2015. PMID: 25785713 Free PMC article.
References
- Blackwell JM. Parasite genome analysis. Progress in the Leishmania genome project. Trans R Soc Trop Med Hyg. 1997;91:107–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U41 HG000739/HG/NHGRI NIH HHS/United States
- P41 HG002223/HG/NHGRI NIH HHS/United States
- P41 HG000739/HG/NHGRI NIH HHS/United States
- P41HG02223/HG/NHGRI NIH HHS/United States
- HG00739/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources