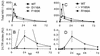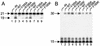Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import - PubMed (original) (raw)
Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import
Ana Limón et al. J Virol. 2002 Nov.
Abstract
Retroviral replication requires the integration of reverse-transcribed viral cDNA into a cell chromosome. A key barrier to forming the integrated provirus is the nuclear envelope, and numerous regions in human immunodeficiency virus type 1 (HIV-1) have been shown to aid the nuclear localization of viral preintegration complexes (PICs) in infected cells. One region in integrase (IN), composed of Val-165 and Arg-166, was reportedly essential for HIV-1 replication and nuclear localization in all cell types. In this study we confirmed that HIV-1(V165A) and HIV-1(R166A) were replication defective and that less mutant viral cDNA localized to infected cell nuclei. However, we present three lines of evidence that argue against a specific role for Val-165 and Arg-166 in PIC nuclear import. First, results of transient transfections revealed that V165A FLAG-tagged IN and green fluorescent protein-IN fusions carrying either V165A or R166A predominantly localized to cell nuclei. Second, two different strains of previously described class II IN mutant viruses displayed similar nuclear entry profiles to those observed for HIV-1(V165A) and HIV-1(R166A), suggesting that defective nuclear import may be a common phenotype of replication-defective IN mutant viruses. Third, V165A and R166A mutants were defective for in vitro integration activity, when assayed both as PICs isolated from infected T-cells and as recombinant IN proteins purified from Escherichia coli. Based on these results, we conclude that HIV-1(V165A) and HIV-1(R166A) are pleiotropic mutants primarily defective for IN catalysis and that Val-165 and Arg-166 do not play a specific role in the nuclear localization of HIV-1 PICs in infected cells.
Figures
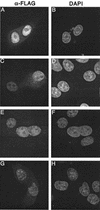
FIG. 1.
FLAG-tagged WT and V165A IN proteins localize to cell nuclei. (A and B) HeLa cells transfected with the codon-optimized WT expression vector. (C and D) Cells transfected with the codon-optimized V165A vector. (E and F) Cells transfected with the previously described (3, 42) FLAG-tagged WT IN expression vector. (G and H) Cells transfected with the previously described V165A vector (3). Fixed cells were mounted in DAPI-containing medium and visualized by indirect immunofluorescence microscopy. Whereas DAPI stained the nuclear DNA of all cells, the anti-FLAG antibody detected IN only in cells that were transfected.
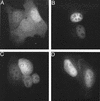
FIG. 2.
Nuclear localization of GFP-IN fusion proteins. (A) HeLa cells were transfected with the pCMX-SAH/Y145F GFP expression vector. (B through D) Cells were transfected with pGFP-IN, pGFP-IN/V165A, and pGFP-IN/R166A expression vectors, respectively. Fixed cells were examined by fluorescence microscopy at 24 h posttransfection.
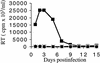
FIG. 3.
HIV-1V165A, HIV-1R166A, and HIV-1R166E are replication- defective in Jurkat T cells. Supernatants of cells infected with WT HIV-1NL4-3 (▪), HIV-1V165A (□), HIV-1R166A (•), HIV-1R166E (○), HIV-1F185K (⧫), or supernatant from mock-transfected cells (×) were assayed for RT activity at the indicated times.
FIG. 4.
HIV-1V165A, HIV-1R166A, HIV-1R166E, and pleiotropic class II IN mutants form similar levels of 2-LTR circles in acute infections. (A) DNA prepared at the indicated times from cells infected with WT HIV-1NL4-3, HIV-1D64N/D116N, HIV-1F185K, or HIV-11-212 was assayed for total HIV-1 content by RQ-PCR. (B) DNA prepared as described in panel A was assayed for 2-LTR circles by RQ-PCR. (C) DNA prepared from cells infected with WT HIV-1NL4-3, HIV-1V165A, HIV-1R166A, or HIV-1R166E was assayed for total HIV-1 cDNA by RQ-PCR. (D) DNA prepared as described in panel C was assayed for 2-LTR circles. Panels B and D are presented with identical _y_-axis ranges for ease of comparing class II and HIV-1V165A, HIV-1R166A, and HIV-1R166E 2-LTR circle levels. Error bars represent variation between duplicate RQ-PCR assays. The class I HIV-1D64N/D116N mutant in panel B formed approximately 21 AU of 2-LTR circles at 24 h. AU, arbitrary units; hpi, hours postinfection.
FIG. 5.
The nuclear localization of HIV-1HXBc2 V165A and R166A mutants resembles the pleiotropic class II F185K mutant. (A) DNA prepared at the indicated times from Jurkat cells infected with WT HIV-1HXBc2, HIV-1D116A, or HIV-1F185K was assayed for total HIV-1 by RQ-PCR. (B) The DNA prepared in panel A was assayed for 2-LTR circles. (C) DNA prepared from cells infected with WT HIV-1HXBc2, HIV-1V165A, or HIV-1R166A was assayed for total HIV-1. (D) DNA prepared in panel C was assayed for 2-LTR circles. HIV-1D116A formed about 1.0 AU of 2-LTR circles at 24 h (panel B). Other labeling is as in Fig. 4.
FIG. 6.
HIV-1V165A and HIV-1R166A do not support detectable levels of in vitro PIC activity. PICs were incubated in the absence or presence of φX174 target DNA as indicated. The lower mobility of the cDNA substrate in lanes 2, 4, and 6 was due to the large excess of target DNA (1.5 μg) present in the integration reaction mixtures. cDNA, 9.7-kb linear HIV-1 substrate; IP, 15.1-kb integration product.
FIG. 7.
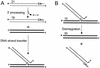
Schematic of in vitro 3′ processing, DNA strand transfer, and disintegration reactions. (A) During 3′ processing, IN cleaves a dinucleotide from the 3′ end of HIV-1 DNA, yielding a product 2 bases shorter than the substrate. During DNA strand transfer, IN joins the processed 3′ end to a 5′ phosphate in target DNA, yielding a population of products longer that the processed DNA strand. (B) Disintegration is essentially a reversal of the DNA strand transfer reaction. Here, IN resolves a branched Y-mer oligonucleotide into separate viral and target DNA duplexes. Thin lines, the U5 end of HIV-1 cDNA; bold lines, target DNA. Numbers refer to lengths of radiolabeled substrate and product DNA strands. ∗, labeled 5′ ends.
FIG. 8.
V165A and R166A IN proteins are defective for 3′ processing and DNA strand transfer activities. (A) 3′ processing activity. IN was omitted from the reaction in lane 1. Lanes 2 to 5, 0.5 μM indicated IN protein; lanes 6 to 9, 0.25 μM IN; 21, migration position of the 21-base labeled strand in substrate DNA; 19, migration position of the cleaved 19-base product (Fig. 7A). (B) Disintegration activity. 15, migration position of the 15-base substrate DNA strand; 30, migration position of the 30-base product (Fig. 7B). Other labeling is as in panel A.
Similar articles
- Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication.
Lu R, Limón A, Devroe E, Silver PA, Cherepanov P, Engelman A. Lu R, et al. J Virol. 2004 Dec;78(23):12735-46. doi: 10.1128/JVI.78.23.12735-12746.2004. J Virol. 2004. PMID: 15542626 Free PMC article. - The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA.
Petit C, Schwartz O, Mammano F. Petit C, et al. J Virol. 2000 Aug;74(15):7119-26. doi: 10.1128/jvi.74.15.7119-7126.2000. J Virol. 2000. PMID: 10888652 Free PMC article. - Nuclear import of the preintegration complex is blocked upon infection by human immunodeficiency virus type 1 in mouse cells.
Tsurutani N, Yasuda J, Yamamoto N, Choi BI, Kadoki M, Iwakura Y. Tsurutani N, et al. J Virol. 2007 Jan;81(2):677-88. doi: 10.1128/JVI.00870-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079325 Free PMC article. - Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV?
Piller SC, Caly L, Jans DA. Piller SC, et al. Curr Drug Targets. 2003 Jul;4(5):409-29. doi: 10.2174/1389450033490984. Curr Drug Targets. 2003. PMID: 12816349 Review. - Nuclear import of human immunodeficiency virus type-1 preintegration complexes.
Fouchier RA, Malim MH. Fouchier RA, et al. Adv Virus Res. 1999;52:275-99. doi: 10.1016/s0065-3527(08)60302-4. Adv Virus Res. 1999. PMID: 10384238 Review.
Cited by
- Integrase-RNA interactions underscore the critical role of integrase in HIV-1 virion morphogenesis.
Elliott JL, Eschbach JE, Koneru PC, Li W, Puray-Chavez M, Townsend D, Lawson DQ, Engelman AN, Kvaratskhelia M, Kutluay SB. Elliott JL, et al. Elife. 2020 Sep 22;9:e54311. doi: 10.7554/eLife.54311. Elife. 2020. PMID: 32960169 Free PMC article. - Lys-34, dispensable for integrase catalysis, is required for preintegration complex function and human immunodeficiency virus type 1 replication.
Lu R, Vandegraaff N, Cherepanov P, Engelman A. Lu R, et al. J Virol. 2005 Oct;79(19):12584-91. doi: 10.1128/JVI.79.19.12584-12591.2005. J Virol. 2005. PMID: 16160186 Free PMC article. - Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses.
Meehan AM, Poeschla EM. Meehan AM, et al. Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):182-91. doi: 10.1016/j.bbagrm.2009.10.001. Epub 2009 Oct 15. Biochim Biophys Acta. 2010. PMID: 19836475 Free PMC article. Review. - PF74 Inhibits HIV-1 Integration by Altering the Composition of the Preintegration Complex.
Balasubramaniam M, Zhou J, Addai A, Martinez P, Pandhare J, Aiken C, Dash C. Balasubramaniam M, et al. J Virol. 2019 Mar 5;93(6):e01741-18. doi: 10.1128/JVI.01741-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567984 Free PMC article. - Nucleocapsid protein function in early infection processes.
Thomas JA, Gorelick RJ. Thomas JA, et al. Virus Res. 2008 Jun;134(1-2):39-63. doi: 10.1016/j.virusres.2007.12.006. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279991 Free PMC article. Review.
References
- Ansari-Lari, M. L., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. - PubMed
- Bouyac-Bertoia M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. - PubMed
- Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI028691/AI/NIAID NIH HHS/United States
- R37 AI039394/AI/NIAID NIH HHS/United States
- R01 AI039394/AI/NIAID NIH HHS/United States
- R01 GM036373/GM/NIGMS NIH HHS/United States
- AI45313/AI/NIAID NIH HHS/United States
- AI39394/AI/NIAID NIH HHS/United States
- AI28691/AI/NIAID NIH HHS/United States
- GM36373/GM/NIGMS NIH HHS/United States
- R37 GM036373/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources