Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence - PubMed (original) (raw)
Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence
Olivier van Eyll et al. J Virol. 2002 Nov.
Abstract
Theiler's virus is a neurotropic murine picornavirus which, depending on the strain, causes either acute encephalitis or persistent demyelinating disease. Persistent strains of Theiler's virus (such as DA) produce an 18-kDa protein called L* from an open reading frame overlapping that encoding the viral polyprotein. Neurovirulent strains (such as GDVII) are thought not to produce the L* protein, as the alternative open reading frame of these strains starts with an ACG codon instead of an AUG codon. However, we observed that both persistent and neurovirulent strain derivatives can produce two forms of the L* protein through unusual type II internal ribosome entry site-mediated translation. A full-length 18-kDa protein can be expressed from an ACG or an AUG initiation codon, whereas an N-terminally truncated 15-kDa product can be translated from a downstream AUG initiation codon. The expression of the 18-kDa form is required for efficient persistence of DA virus derivatives in the central nervous system.
Figures
FIG. 1.
L* mutant viruses. The nucleotide and amino acid sequences of L* mutations are shown. The various mutations are indicated under the corresponding sequence segments of the parental strain. Mutated nucleotides are underlined. Nucleotides of the main ORF upstream of the L* ORF are in lowercase. Numbering corresponds to the codons of the L* protein. The nucleotide sequence of the neurovirulent genome is indicated in bold. For each parental strain, a table summarizes the mutations present in the corresponding mutant viruses. (A) DA1 derivatives. (B) GDVII and OV47. (C) R2 and OV57. Note that OV23 and OV48 are identical. OV48 was derived from OV46 to confirm data obtained with OV23.
FIG. 2.
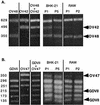
Detection of viral RNA in the brains and spinal cords of infected SJL mice 45 days postinoculation. Viral RNA was detected by dot blot hybridization of total RNA extracted from the brains and spinal cords of four mice inoculated with 105 PFU of the indicated viruses. Control mice (T−) were mock infected. Viruses and corresponding L* ORF mutations are shown to the left and right, respectively.
FIG. 3.
Mixed infections of fibroblast and macrophage cell lines. A mixture of two viruses (1:1) was prepared and used to infect BHK-21 and RAW264.7 (RAW) cells. RNA was extracted from infected cells after one to five passages of the virus mixture. As a control, RNA was extracted from the virus mixture before infection and from the parental virus stocks. The L* region was amplified by RT-PCR and digested with a restriction enzyme, allowing discrimination between the two viruses present in the mixture. (Left panels) Control analysis of the parental viruses and of the mixture used. (Right panels) Analysis of the virus mixture contained in the infected cells after one (P1) to five (P5) passages. Arrowheads indicate the fragments that are diagnostic of a given virus. (A) Competition between OV48, an AUG-to-ACG mutant, and OV42, a stop codon 93 mutant of DA1. _Tru_I was used to digest the PCR fragments. The sizes of the fragments are indicated in base pairs on the left. (B) Competition between GDVII and its stop codon derivative, OV47. _Hsp_92II was used to digest the PCR fragments.
FIG. 4.
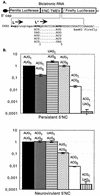
Bicistronic constructs and IRES-mediated expression of luciferase from AUG or ACG initiation codons. (A) Bicistronic constructs in which translation of the firefly luciferase gene is under the control of the TMEV IRES. Constructs were made in the pHMG vector background (15). The 5′ end of the L* ORF, followed by a _Bam_HI restriction site, is fused in frame with the second codon of the firefly luciferase gene. The polyprotein and L* AUG initiation codons are shown in bold. The Kozak context sequence of the first L* AUG is underlined. The sequence of the firefly luciferase gene is italicized. The different codon combinations replacing AUG1 and AUG5 of L* are shown under the sequence. NC, noncoding region. Nucleotides of the L*-luciferase gene fusion are in uppercase. Nucleotides of the main ORF upstream of the L* ORF are in lowercase. (B) Luciferase activity expressed by the different constructs in BHK-21 cells. The bicistronic constructs shown in panel A were transfected into BHK-21 cells. At 24 h posttransfection, the level of expression of both luciferase genes was measured. The firefly/Renilla luciferase expression ratio (construct:control) was calculated and normalized to the value obtained for the AUG1-AUG5 combination. The histograms show means and standard deviations for at least three transfection experiments.
FIG. 5.
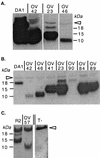
Detection by Western blotting of L* protein expression in infected BHK-21 cells. (A) Comparison of the amounts of L* protein produced by viruses with AUG or ACG: wild type virus DA1 and mutant viruses OV23, OV42, and OV46. (B) Comparison of the viruses in panel A and OV41, OV84, OV89, and OV90 mutant viruses. Note that samples from DA1- and OV42-infected cells were diluted 100-fold to allow a better molecular mass comparison. (C) Comparison of recombinant virus R2 and the corresponding stop codon mutant virus, OV57. The arrowhead points to a nonspecific band detected in both infected and mock-infected cell extracts. T-, mock-infected cell extract.
FIG. 6.
L* protein expression and viral persistence. The expected L* protein products for the various L* mutants are represented by bars. The black and gray bars represent the L* protein products initiated from AUG and ACG codons, respectively. An asterisk symbolizes the Met41-to-Thr mutation of the L* protein introduced to mutate the AUG41 codon. For each L* product, the expected molecular mass (L* prot. kDa) and the estimated amount of protein detected by Western blotting in BHK-21 cells (L* expr. W-B) are shown. +++++, wild-type expression level; +++, intermediate expression level; +, low expression level; −, not detectable; n.d., not determined. The persistence of the various mutants in the CNS, as deduced from dot blot (Fig. 2) or real-time PCR (Table 1) measurements, is indicated (Persist. CNS).
FIG. 7.
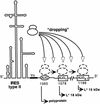
Dropping model for type II IRES-mediated multiple translation initiation. Yn, oligopyrimidine tract conserved in picornavirus IRESs. The circled AUG is the start codon for translation of the polyprotein. Boxed AUG codons are codons that would initiate translation of the 18- and 15-kDa L* products. Instead of positioning the small ribosomal subunit at a fixed position corresponding to the initiation codon, the IRES would be more “flexible” and drop the ribosome at various positions. From there, the ribosome could scan to find the first available initiator codon.
Similar articles
- Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence.
van Eyll O, Michiels T. van Eyll O, et al. J Virol. 2000 Oct;74(19):9071-7. doi: 10.1128/jvi.74.19.9071-9077.2000. J Virol. 2000. PMID: 10982352 Free PMC article. - Alternative translation initiation of Theiler's murine encephalomyelitis virus.
Yamasaki K, Weihl CC, Roos RP. Yamasaki K, et al. J Virol. 1999 Oct;73(10):8519-26. doi: 10.1128/JVI.73.10.8519-8526.1999. J Virol. 1999. PMID: 10482605 Free PMC article. - Theiler's murine encephalomyelitis virus L* amino acid position 93 is important for virus persistence and virus-induced demyelination.
Stavrou S, Baida G, Viktorova E, Ghadge G, Agol VI, Roos RP. Stavrou S, et al. J Virol. 2010 Feb;84(3):1348-54. doi: 10.1128/JVI.01585-09. Epub 2009 Nov 18. J Virol. 2010. PMID: 19923182 Free PMC article. - The genetics of the persistent infection and demyelinating disease caused by Theiler's virus.
Brahic M, Bureau JF, Michiels T. Brahic M, et al. Annu Rev Microbiol. 2005;59:279-98. doi: 10.1146/annurev.micro.59.030804.121242. Annu Rev Microbiol. 2005. PMID: 16153171 Review.
Cited by
- Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4.
McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. McFadden N, et al. PLoS Pathog. 2011 Dec;7(12):e1002413. doi: 10.1371/journal.ppat.1002413. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174679 Free PMC article. - Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein.
Ricour C, Delhaye S, Hato SV, Olenyik TD, Michel B, van Kuppeveld FJ, Gustin KE, Michiels T. Ricour C, et al. J Gen Virol. 2009 Jan;90(Pt 1):177-86. doi: 10.1099/vir.0.005678-0. J Gen Virol. 2009. PMID: 19088287 Free PMC article. - Identification of a novel neuropathogenic Theiler's murine encephalomyelitis virus.
Buckwalter MR, Nga PT, Gouilh MA, Fiette L, Bureau JF, Laird ME, Buchrieser J, Ozden S, Cheval J, Eloit M, Manuguerra JC, Gessain A, Brey PT, Fontanet A, Albert ML. Buckwalter MR, et al. J Virol. 2011 Jul;85(14):6893-905. doi: 10.1128/JVI.00274-11. Epub 2011 May 4. J Virol. 2011. PMID: 21543488 Free PMC article. - Leader (L) and L* proteins of Theiler's murine encephalomyelitis virus (TMEV) and their regulation of the virus' biological activities.
Takano-Maruyama M, Ohara Y, Asakura K, Okuwa T. Takano-Maruyama M, et al. J Neuroinflammation. 2006 Aug 16;3:19. doi: 10.1186/1742-2094-3-19. J Neuroinflammation. 2006. PMID: 16911804 Free PMC article. - Different subcellular localization of Theiler's murine encephalomyelitis virus leader proteins of GDVII and DA strains in BHK-21 cells.
Taniura N, Saito M, Okuwa T, Saito K, Ohara Y. Taniura N, et al. J Virol. 2009 Jul;83(13):6624-30. doi: 10.1128/JVI.02385-08. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386716 Free PMC article.
References
- Aubert, C., M. Chamorro, and M. Brahic. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319-326. - PubMed
- Brahic, M., W. G. Stroop, and J. R. Baringer. 1981. Theiler's virus persists in glial cells during demyelinating disease. Cell 26:123-128. - PubMed
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
- Drescher, K. M., L. R. Pease, and M. Rodriguez. 1997. Antiviral immune responses modulate the nature of central nervous system (CNS) disease in a murine model of multiple sclerosis. Immunol. Rev. 159:177-193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources