Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59 - PubMed (original) (raw)
Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59
Boyd Yount et al. J Virol. 2002 Nov.
Abstract
A novel method was developed to assemble a full-length infectious cDNA of the group II coronavirus mouse hepatitis virus strain A59 (MHV-A59). Seven contiguous cDNA clones that spanned the 31.5-kb MHV genome were isolated. The ends of the cDNAs were engineered with unique junctions and assembled with only the adjacent cDNA subclones, resulting in an intact MHV-A59 cDNA construct of approximately 31.5 kb in length. The interconnecting restriction site junctions that are located at the ends of each cDNA are systematically removed during the assembly of the complete full-length cDNA product, allowing reassembly without the introduction of nucleotide changes. RNA transcripts derived from the full-length MHV-A59 construct were infectious, although transfection frequencies were enhanced 10- to 15-fold in the presence of transcripts encoding the nucleocapsid protein N. Plaque-purified virus derived from the infectious construct replicated efficiently and displayed similar growth kinetics, plaque morphology, and cytopathology in murine cells as did wild-type MHV-A59. Molecularly cloned viruses recognized the MHV receptor (MHVR) for docking and entry, and pretreatment of cells with monoclonal antibodies against MHVR blocked virus entry and replication. Cells infected with molecularly cloned MHV-A59 virus expressed replicase (gene 1) proteins identical to those of laboratory MHV-A59. Importantly, the molecularly cloned viruses contained three marker mutations that had been derived from the engineered component clones. Full-length infectious constructs of MHV-A59 will permit genetic modifications of the entire coronavirus genome, particularly in the replicase gene. The method has the potential to be used to construct viral, microbial, or eukaryotic genomes approaching several million base pairs in length and used to insert restriction sites at any given nucleotide in a microbial genome.
Figures
FIG.1.
Strategy to assemble an infectious cDNA of MHV-A59. (A) _Esp_3I cloning properties. In the traditional mode, an _Esp_3I site resides at the 3′ end of the MHV A subclone and upon cleavage results in a novel 4-nucleotide overhang which will specifically anneal with the complementary 4-nucleotide overhang generated by an identical _Esp_3I site located at the 5′ end of the MHV B fragment. Note that the MHV wild-type sequence is re-formed intact and the _Esp_3I site is retained in the virus sequence. However, the _Esp_3I recognition site is a nonpalindromic sequence, and as such, a simple flip in orientation allows for the specific removal of the _Esp_3I recognition site in the MHV A and B subclones, leaving 4-nucleotide complementary overhangs, which re-form the intact wild-type sequence upon ligation. No evidence of the _Esp_3I site that has been engineered into the component clones should remain in the assembled product (No See'm technology). In this cartoon, the sequences represent wild-type MHV sequence but are not the exact nucleotides encoded at the MHV A/B junction (Tables 1 and 2). (B) Component clones used in the assembly of a full-length MHV-A59 cDNA. The MHV genome is a linear positive-polarity RNA of about 31,500 nucleotides in length. By using RT-PCR and unique oligonucleotide primer mutagenesis, seven clones spanning the entire MHV genome were isolated by standard recombinant DNA techniques. Unique _Esp_3I and _Bgl_I sites, located at the junctions between each of the cDNA clones, were used to assemble a full-length infectious cDNA. A unique T7 start site was inserted at the 5′ end of clone A, and a 25-nucleotide T tail and downstream _Sfi_I site were encoded at the 3′ end of clone G. The approximate location of each site is shown. (C) Location of the MHV component clone junctions. MHV ORF1a and ORF1b polyproteins are proteolytically processed into numerous smaller proteins, which presumably function to direct virus replication and transcription. The component clone A/B, B/C, C/D, D/E, and E/F junctions are shown relative to the ORF1 polyproteins synthesized during MHV-A59 infection. The F/G junction resides in the HE glycoprotein pseudogene. utr, untranslated region; nt, nucleotide.
FIG. 2.
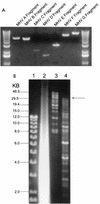
Assembly of an MHV-A59 full-length cDNA. (A) Plasmid DNAs were restricted with the appropriate enzymes and separated in 0.8% agarose gels, and the MHV cDNAs were purified from gels. The sizes (in kilobases) of the MHV fragments are as follows: A, ∼4.8; B, ∼4.3; C, ∼2.0; D, ∼1.5; E, ∼2.8; F, ∼7.0; and G, ∼8.7. DNA molecular size markers flank the MHV cDNAs. (B) The purified cDNAs were pooled and incubated with T4 DNA ligase for 12 h at 4°C. One-thirtieth of the assembled products were separated in 0.5% agarose gels overnight at 15 V. Lane 1, small-molecular-size 1-kb markers; lane 2, in vitro transcripts derived from MHV-A59 1000 and treated with DNase I; lane 3, high-molecular-weight markers; lane 4, shotgun-assembled MHV-A59 component cDNAs. The arrow indicates the presence of large-molecular-size ∼31.5-kb cDNA.
FIG. 3.
Molecularly cloned virus plaque morphology. Cultures of BHK cells were electroporated with MHV-A59 1000 full-length transcripts or MHV-A59 1000 cDNA and seeded with DBT cells in 75-cm2 flasks. Virus progeny were purified by plaque assay, and virus stocks were grown as described in Materials and Methods. The figure shows the plaque morphologies of wild-type MHV-A59 virus (A) and molecularly cloned icMHV-A59#1 (B) and icMHV-A59#2 (C) virus.
FIG. 4.
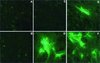
Viral antigen expression in the presence or absence of nucleocapsid transcripts. Cultures of BHK-MHVR cells were electroporated with MHV-A59 1000 alone or with a mixture of MHV-A59 1000 and N gene transcripts as described in Materials and Methods. In LabTek chambers, 2 × 105 cells were seeded and fixed at 16 and 25 h posttransfection. With murine antiserum against MHV-A59, fluorescent antibody staining demonstrated that MHV-A59 1000 transcripts were infectious in the presence or absence of N transcripts. (A B) Untransfected culture at 16 h; (B) untransfected culture at 25 h; (C) culture transfected in the absence of N transcripts, at 16 h; (C) culture transfected in the presence of N transcripts, at 16 h; (E) culture transfected in the absence of N transcripts, at 25 h (F) culture transfected in the presence of N transcripts, at 25 h.
FIG. 5.
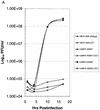
Growth curves of molecularly cloned and wild-type MHV-A59. Cultures of 2.0 × 105 DBT, BHK, or BHK-MHVR cells were infected with various molecularly cloned viruses and wild-type MHV-A59 at an MOI of 10 for 1 h. In some instances prior to infection, the cells had been pretreated with a 1:2 dilution of monoclonal antibody CC1, directed against the MHVR. Virus samples were taken at the indicated times. The figure shows virus growth in DBT (A), BHK-MHVR (B), and BHK (C) cells, respectively.
FIG. 5.
Growth curves of molecularly cloned and wild-type MHV-A59. Cultures of 2.0 × 105 DBT, BHK, or BHK-MHVR cells were infected with various molecularly cloned viruses and wild-type MHV-A59 at an MOI of 10 for 1 h. In some instances prior to infection, the cells had been pretreated with a 1:2 dilution of monoclonal antibody CC1, directed against the MHVR. Virus samples were taken at the indicated times. The figure shows virus growth in DBT (A), BHK-MHVR (B), and BHK (C) cells, respectively.
FIG. 5.
Growth curves of molecularly cloned and wild-type MHV-A59. Cultures of 2.0 × 105 DBT, BHK, or BHK-MHVR cells were infected with various molecularly cloned viruses and wild-type MHV-A59 at an MOI of 10 for 1 h. In some instances prior to infection, the cells had been pretreated with a 1:2 dilution of monoclonal antibody CC1, directed against the MHVR. Virus samples were taken at the indicated times. The figure shows virus growth in DBT (A), BHK-MHVR (B), and BHK (C) cells, respectively.
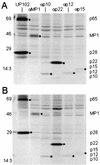
FIG. 6.
Expression of replicase proteins of wild-type MHV-A59 and icMHV-A59. Wild-type MHV-A59 and icMHV-A59#1 were inoculated onto monolayers of DBT cells at an MOI of 10 for 1 h. Cells were maintained for 5.5 h in Dulbecco modified Eagle medium-2% fetal calf serum, and at 5.5 h, the media were replaced with Dulbecco modified Eagle medium that lacked methionine and cysteine and contained 2% fetal calf serum. Actinomycin D (2 μg/ml) was also added at 5.5 h p.i., and at 6.0 h, the cultures were pulsed with [35S]Met for 2 h. Cells were lysed at 8 h p.i. in lysis buffer containing 10 mM Tris-HCl, 1% NP-40, and 2% SDS. Immunoprecipitation of replicase proteins was performed in 200-μl reaction mixtures containing lysate from ∼105 cells in 10 mM Tris-1% NP-40-1% SDS. The specificities and sensitivities of the antibodies directed against p28/p65 (UP102), MP1 (αMP1), 3CLpro (B3), p12 (αp1a-12), p22 (αp22), p10 (αp1a-10), and p15 (αp1a-15) have been reported previously (10, 19, 20, 31, 32). Molecular weight markers are shown to the left of each gel, and black circles identify the various MHV replicase protein products.
FIG. 7.
Restriction fragment length polymorphism analysis of molecularly cloned virus. Cultures were infected with MHV-A59 or icMHV-A59#1, and intracellular RNA was isolated at 8 h p.i. With primer pairs and RT-PCR, cDNA amplicons were isolated that contained the various marker mutations that had been inserted into the component clones. The purified wild-type MHV-A59 and icMHV-A59#1 amplicons were restricted with _Esp_3I or _Rsr_II and separated in 0.8% agarose gels. Lane 1, wild-type A59 amplicon from nucleotides 2020 to 5031, uncut; lane 2, wild-type A59 amplicon from nucleotides 2020 to 5031 digested with _Esp_3I; lane 3, icMHV-A59#1 amplicon from nucleotides 2020 to 5031, uncut; lane 4, icMHV-A59#1 amplicon from nucleotides 2020 to 5031, _Esp_3I digested; lane 5, wild-type A59 amplicon from nucleotides 16351 to 17875, uncut; lane 6, wild-type A59 amplicon from nucleotides 16351 to 17875 restricted with _Esp_3I; lane 7, icMHV-A59#1 amplicon from nucleotides 16351 to 17875, uncut; lane 8, icMHV-A59#1 amplicon from nucleotides 16351 to 17875 restricted with _Esp_3I; lane 9, wild-type A59 amplicon from nucleotides 22060 to 25416, uncut; lane 10, wild-type A59 amplicon from nucleotides 22060 to 25416 restricted with _Rsr_II; lane 11, icMHV-A59#1 amplicon from nucleotides 22060 to 25416, uncut; lane 12, icMHV-A59#1 amplicon from nucleotides 22060 to 25416 restricted with _Rsr_II; lane 13, 1-kb ladder.
FIG. 8.
Identification of marker mutations in recombinant virus. Various icMHV-A59#1 or wild-type virus amplicons were subcloned into Topo II vectors and sequenced. (A) icMHV-A59#1 sequence across the mutated 3512 _Esp_3I site; (B) icMHV-A59#1 sequence across the mutated 48 _Esp_3I site and the MHV A/B junction; (C) icMHV-A59#1 sequence across the mutated 17 _Esp_3I site; (D) icMHV-A59#1 sequence across the _Rsr_II site and MHV F/G junction; (E) wild-type MHV-A59 sequence across the same domain as shown in panel D. Overlined sequences include the mutated restriction sites and the corresponding wild-type nucleotides at these positions. In panels B and D, the bracketed sequences represent the MHV A/B and F/G junction domains, respectively. Note that all evidence of the _Esp_3I site, which had been engineered into the sequence, is gone.
Similar articles
- The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range.
Schickli JH, Zelus BD, Wentworth DE, Sawicki SG, Holmes KV. Schickli JH, et al. J Virol. 1997 Dec;71(12):9499-507. doi: 10.1128/JVI.71.12.9499-9507.1997. J Virol. 1997. PMID: 9371612 Free PMC article. - Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model.
Yount B, Curtis KM, Baric RS. Yount B, et al. J Virol. 2000 Nov;74(22):10600-11. doi: 10.1128/jvi.74.22.10600-10611.2000. J Virol. 2000. PMID: 11044104 Free PMC article. - Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice.
Sperry SM, Kazi L, Graham RL, Baric RS, Weiss SR, Denison MR. Sperry SM, et al. J Virol. 2005 Mar;79(6):3391-400. doi: 10.1128/JVI.79.6.3391-3400.2005. J Virol. 2005. PMID: 15731233 Free PMC article. - Selection in persistently infected murine cells of an MHV-A59 variant with extended host range.
Schickli JH, Wentworth DE, Zelus BD, Holmes KV, Sawicki SG. Schickli JH, et al. Adv Exp Med Biol. 1998;440:735-41. doi: 10.1007/978-1-4615-5331-1_95. Adv Exp Med Biol. 1998. PMID: 9782352 - Development of mouse hepatitis virus and SARS-CoV infectious cDNA constructs.
Baric RS, Sims AC. Baric RS, et al. Curr Top Microbiol Immunol. 2005;287:229-52. doi: 10.1007/3-540-26765-4_8. Curr Top Microbiol Immunol. 2005. PMID: 15609514 Free PMC article. Review.
Cited by
- Characterization of a pathogenic full-length cDNA clone of a virulent porcine epidemic diarrhea virus strain AH2012/12 in China.
Fan B, Yu Z, Pang F, Xu X, Zhang B, Guo R, He K, Li B. Fan B, et al. Virology. 2017 Jan;500:50-61. doi: 10.1016/j.virol.2016.10.011. Epub 2016 Oct 20. Virology. 2017. PMID: 27770703 Free PMC article. - SARS CoV replication and pathogenesis in human airway epithelial cultures.
Sims AC, Yount B, Burkett SE, Baric RS, Pickles RJ. Sims AC, et al. Adv Exp Med Biol. 2006;581:535-8. doi: 10.1007/978-0-387-33012-9_97. Adv Exp Med Biol. 2006. PMID: 17037593 Free PMC article. No abstract available. - Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function.
Gadlage MJ, Sparks JS, Beachboard DC, Cox RG, Doyle JD, Stobart CC, Denison MR. Gadlage MJ, et al. J Virol. 2010 Jan;84(1):280-90. doi: 10.1128/JVI.01772-09. J Virol. 2010. PMID: 19846526 Free PMC article. - The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication.
Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. Graham RL, et al. J Virol. 2005 Nov;79(21):13399-411. doi: 10.1128/JVI.79.21.13399-13411.2005. J Virol. 2005. PMID: 16227261 Free PMC article. - Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone.
St-Jean JR, Desforges M, Almazán F, Jacomy H, Enjuanes L, Talbot PJ. St-Jean JR, et al. J Virol. 2006 Apr;80(7):3670-4. doi: 10.1128/JVI.80.7.3670-3674.2006. J Virol. 2006. PMID: 16537637 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI026603/AI/NIAID NIH HHS/United States
- GM63228/GM/NIGMS NIH HHS/United States
- AI17418/AI/NIAID NIH HHS/United States
- R01 AI017418/AI/NIAID NIH HHS/United States
- R01 AI023946/AI/NIAID NIH HHS/United States
- AI23946/AI/NIAID NIH HHS/United States
- R01 GM063228/GM/NIGMS NIH HHS/United States
- AI26603/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources