Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element - PubMed (original) (raw)
Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element
Takeshi Kuwata et al. Mol Cell Biol. 2002 Nov.
Abstract
ICSBP (IRF-8) is a transcription factor of the IRF family expressed only in the immune system. It is induced in macrophages by gamma interferon (IFN-gamma) and contributes to macrophage functions. By interacting with Ets family protein PU.1, ICSBP binds to the IRF/Ets composite element and stimulates transcription. ICSBP binds to another DNA element, the IFN-stimulated response element (ISRE), a common target of the IRF family. Limited knowledge as to how ICSBP and other IRF proteins regulate ISRE-dependent transcription in IFN-gamma-activated macrophages is available. By mass-spectrometric analysis of ISRE-bound proteins in macrophages, we identified TEL, another Ets member, as a factor recruited to the element in an IFN-gamma-dependent manner. In vitro analysis with recombinant proteins indicated that this recruitment is due to a direct interaction between ICSBP and TEL, which is enhanced by the presence of ISRE. Significantly, the interaction with TEL in turn resulted in the recruitment of the histone deacetytase HDAC3 to the ISRE, causing increased repression of IFN-gamma-mediated reporter activity through the ISRE. This repression may provide a negative-feedback mechanism operating after the initial transcriptional activation by IFN-gamma. By associating with two different Ets family proteins, ICSBP exerts a dual function in IFN-gamma-dependent gene regulation in an immune system-specific manner.
Figures
FIG. 1.
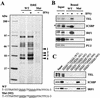
IFN-γ-dependent recruitment of TEL to the ISRE. (A) Three copies of the wild-type (WT) and mutant (Mut) ISRE immobilized to magnetic beads were incubated with nuclear extracts from RAW cells treated with or without IFN-γ (100 U/ml) for 12 h. Bound materials were separated by SDS-8% PAGE and visualized by silver staining. Bands 1 and 2 and band 3 (arrowheads) were identified as TEL and ICSBP, respectively (Table 1). (B) Immobilized wild-type ISREs were incubated with 100 μg of extracts from RAW cells treated with IFN-γ for 12 h, and bound proteins were separated by SDS-10% PAGE and detected by immunoblotting. Input represents 5% of total extracts. ∗, nonspecific protein that did not bind to the ISRE. (C) The wild-type and mutant ISREs were immobilized to the beads, and the binding of the indicated factors was tested by immunoblotting as for panel B. Input represents 5% of total extracts.
FIG. 2.
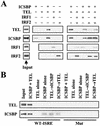
ICSBP-dependent recruitment of recombinant TEL to the ISRE. (A) Immobilized wild-type ISREs were incubated with 20 ng of the indicated recombinant proteins for 1 h at 4°C, and bound proteins were detected by immunoblot analysis. Input represents 10% of the total reaction mixture. (B) Immobilized wild-type (WT) and mutant (Mut) ISREs (M2) were first incubated with 50 ng of the indicated recombinant proteins for 1 h at 4°C, washed, and then incubated with the second protein under the same conditions. Bound proteins were detected as for panel A. Input represents 5% of total extracts.
FIG. 3.
Domains required for TEL-ICSBP interaction. (A) (Top) Diagram of TEL deletion mutants and summary of ICSBP binding. (Bottom) Coomassie blue staining of GST-TEL fusions and binding of 35S-labeled ICSBP. Input represents 10% of total reaction. WT, wild type. (B) (Top) Diagram of ICSBP deletion mutants and summary of TEL binding. (Bottom) 35S-lableled ICSBP deletion mutants were incubated with control GST or GST-full-length TEL, and bound materials were detected by autoradiography. Input represents 10% of the reaction mixture.
FIG. 4.
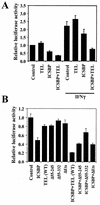
Increased repression of ISRE-reporter activity by TEL and ICSBP. (A) RAW cells were transfected with 0.4 μg of ISRE-luciferase reporter along with 0.4 μg of pcx-ICSBP and/or pcDNA-TEL and the Renilla plasmid (5 ng) for 12 h. The total amount of transfected DNA was adjusted with appropriate empty vector to 1.205 μg. Cells were then treated with or without IFN-γ for 12 h. Luciferase activity was normalized by the Renilla internal control. Values represent the averages of three determinations ± standard deviations (SD). (B) RAW cells were transfected with ISRE-reporter along with pcx-ICSBP and the indicated TEL deletion mutants and the Renilla control as for panel A. Cells were treated with IFN-γ for 12 h, and luciferase activity was measured as for panel A. Values represent the averages of three determinations ± SD.
FIG. 5.
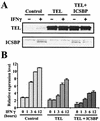
Reduced 2′,5′-OAS transcript expression by TEL and ICSBP. (A) 32D cells were stably transfected with the control vector or full-length TEL, and appropriate clones were isolated. The TEL clone was transduced with control (TEL) or ICSBP retrovirus vector (TEL + ICSBP). Cells were treated with IFN-γ (100 U/ml) for 6 h. Expression of TEL and ICSBP proteins was detected by immunoblot analysis using 100 μg of nuclear extracts. (B) 2′,5′-OAS mRNA levels in indicated 32D clones treated with IFN-γ (100 U/ml) for the indicated periods were measured by real-time reverse transcription-PCR. Values represent relative mRNA levels normalized by HPRT.
FIG. 6.
IFN-γ-dependent recruitment of HDAC3 to the ISRE. (A and B) The immobilized wild-type (WT) and mutant (Mut) ISREs were incubated with 100 μg of extracts from RAW cells treated with IFN-γ (100 U/ml) (A and B) or IFN-α/β (1,000 U/ml) (B) for 12 h, and bound proteins were detected by immunoblotting. Input represents 5% of total extracts. (C) Binding of indicated factors was tested with 100 μg of extracts from RAW cells treated for the indicated times. Input represents 10% of total extracts.
FIG. 7.
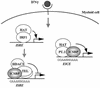
A model for interaction of ICSBP with TEL. ICSBP interacts with Ets family proteins TEL and PU.1 in IFN-γ-stimulated macrophages. The interaction with TEL occurs on the ISRE, while that with PU.1 requires the EICE. The two partners bind to the elements in opposite orientations: on the ISRE, ICSBP binds to the 5′ site and TEL binds to the 3′ site, while on the EICE, PU.1 binds the 5′ site and ICSBP binds to the 3′ site. ICSBP-TEL complex formation may be a late event that occurs following transcriptional activation by IRF-1, as it recruits HDAC3 and represses ISRE-dependent transcription. This repression may be part of a negative-feedback mechanism. On the other hand, the ICSBP-PU.1 complex activates EICE-dependent transcription presumably by recruiting the histone acetylase (HAT) CBP/p300. This model illustrates a dual role of ICSBP that is acquired by the interaction with two different Ets family partners, permitting immune cell-specific gene regulation by IFN-γ.
Similar articles
- Complex formation of the interferon (IFN) consensus sequence-binding protein with IRF-1 is essential for murine macrophage IFN-gamma-induced iNOS gene expression.
Xiong H, Zhu C, Li H, Chen F, Mayer L, Ozato K, Unkeless JC, Plevy SE. Xiong H, et al. J Biol Chem. 2003 Jan 24;278(4):2271-7. doi: 10.1074/jbc.M209583200. Epub 2002 Nov 11. J Biol Chem. 2003. PMID: 12429737 - Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex.
Kanno Y, Levi BZ, Tamura T, Ozato K. Kanno Y, et al. J Interferon Cytokine Res. 2005 Dec;25(12):770-9. doi: 10.1089/jir.2005.25.770. J Interferon Cytokine Res. 2005. PMID: 16375605 Review. - ICSBP/IRF-8 transactivation: a tale of protein-protein interaction.
Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A, Meraro D. Levi BZ, et al. J Interferon Cytokine Res. 2002 Jan;22(1):153-60. doi: 10.1089/107999002753452764. J Interferon Cytokine Res. 2002. PMID: 11846986 Review.
Cited by
- IRF8 regulates myeloid and B lymphoid lineage diversification.
Wang H, Morse HC 3rd. Wang H, et al. Immunol Res. 2009;43(1-3):109-17. doi: 10.1007/s12026-008-8055-8. Immunol Res. 2009. PMID: 18806934 Free PMC article. Review. - Transcription-independent regulation of STING activation and innate immune responses by IRF8 in monocytes.
Luo WW, Tong Z, Cao P, Wang FB, Liu Y, Zheng ZQ, Wang SY, Li S, Wang YY. Luo WW, et al. Nat Commun. 2022 Aug 16;13(1):4822. doi: 10.1038/s41467-022-32401-1. Nat Commun. 2022. PMID: 35973990 Free PMC article. - Single-cell chromatin accessibility and transcriptomic characterization of Behcet's disease.
Shi W, Ye J, Shi Z, Pan C, Zhang Q, Lin Y, Liang D, Liu Y, Lin X, Zheng Y. Shi W, et al. Commun Biol. 2023 Oct 17;6(1):1048. doi: 10.1038/s42003-023-05420-x. Commun Biol. 2023. PMID: 37848613 Free PMC article. - Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages.
Tamura T, Thotakura P, Tanaka TS, Ko MS, Ozato K. Tamura T, et al. Blood. 2005 Sep 15;106(6):1938-47. doi: 10.1182/blood-2005-01-0080. Epub 2005 Jun 9. Blood. 2005. PMID: 15947094 Free PMC article. - IFN-γ production by brain-resident cells activates cerebral mRNA expression of a wide spectrum of molecules critical for both innate and T cell-mediated protective immunity to control reactivation of chronic infection with Toxoplasma gondii.
Suzuki Y, Lutshumba J, Chen KC, Abdelaziz MH, Sa Q, Ochiai E. Suzuki Y, et al. Front Cell Infect Microbiol. 2023 Feb 15;13:1110508. doi: 10.3389/fcimb.2023.1110508. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36875520 Free PMC article.
References
- Bergman, A. C., T. Benjamin, A. Alaiya, M. Waltham, K. Sakaguchi, B. Franzen, S. Linder, T. Bergman, G. Auer, E. Appella, P. J. Wirth, and H. Jornvall. 2000. Identification of gel-separated tumor marker proteins by mass spectrometry. Electrophoresis 21:679-686. - PubMed
- Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. - PubMed
- Brass, A. L., E. Kehrli, C. F. Eisenbeis, U. Storb, and H. Singh. 1996. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10:2335-2347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources