Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase - PubMed (original) (raw)
Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase
Xing Zhang et al. Cell. 2002.
Abstract
AdoMet-dependent methylation of histones is part of the "histone code" that can profoundly influence gene expression. We describe the crystal structure of Neurospora DIM-5, a histone H3 lysine 9 methyltranferase (HKMT), determined at 1.98 A resolution, as well as results of biochemical characterization and site-directed mutagenesis of key residues. This SET domain protein bears no structural similarity to previously characterized AdoMet-dependent methyltransferases but includes notable features such as a triangular Zn3Cys9 zinc cluster in the pre-SET domain and a AdoMet binding site in the SET domain essential for methyl transfer. The structure suggests a mechanism for the methylation reaction and provides the structural basis for functional characterization of the HKMT family and the SET domain.
Figures
Figure 1. Structure-Based Sequence Alignment of SET Proteins
The alignment includes (1) all known members of human SUV39 family: SUV39H1 (NP_003164), SUV39H2 (NP_078946), G9a (S30385), Eu-HMT1 (AAM09024), SETDB1 (NP_036564), and CLLL8 (NP_114121); (2) proteins (in bold) that have been shown to have HKMT activity from various species: N. crassa (Nc) DIM-5 (AAL35215), S. pombe (Sp) Clr4 (060016), A. thaliana (At) SUVH4 or KRYPTONITE (AAK28969), S. cerevisiae (Sc) SET1 (P38827) and SET2 (P46995), human SET7 (XP_040150) and PR-SET7 (AAL40879); (3) three bacterial SET proteins: Xylella fastidiosa (Xf) SET (AAF84287), Bradyrhizobium japonicum (Bj) SET (Q9ANB6), and Chlamydophila pneumoniae (Cp) SET (AAD19016); and (4) human EZH2 protein, which appears inactive in vitro (Rea et al., 2000). The residue number and secondary structural elements of DIM-5 (helices A–J and strands 1–17) are shown above the aligned sequences. Dashed lines indicate disordered regions. The color coding is light blue for the N terminus (residues 25–62), yellow for the pre-SET (residues 63–146), green for the SET (residues 147–236 and 248–277), magenta for the signature motifs (SET residues 237–247 and 278–285), and gray for the post-SET (residues 299–308). The amino acids highlighted are invariant (white against black) and conserved (white against gray) among almost all members of the SUV39 family. The number in parentheses indicates the number of amino acids inserted relative to the alignment. The lowercase letters above the sequences indicate the structural/functional role of the corresponding DIM-5 residues: “h” indicates intramolecular hydrophobic interaction, “n” indicates intramolecular nonhydrophobic (polar or charge) interaction, “z” indicates zinc coordination, asterisk indicates structural residue Gly or Pro, and “s” indicates surface-exposed residues potentially important for cofactor or substrate binding or catalysis. The red circles mark the residues that were mutated in this study.
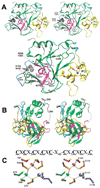
Figure 2. DIM-5 Structure
(A) Front view of ribbons diagram (Carson, 1997) (top, stereo; bottom, mono). The protein is colored according to Figure 1, and the three zinc ions are red, green, and blue balls (as in C). The difference electron density map (black), contoured at 5.5σ above the mean, indicates the presumed cofactor binding site (supported by tests on mutant forms). (B) Side view. A dashed line indicates the disordered amino acids between strand β17 (magenta) and the post-SET segment (gray). (C) Stereo diagram of the triangular zinc cluster. Three zinc ions are colored in red, green, and blue, the bridging cysteine residues are colored in orange, and the nonbridging cysteine residues are colored to match their associated zinc ion. The atoms are gray (carbon) and yellow (sulfur). The pre-SET sequence of DIM-5 is shown above. Both Cys-rich segments coordinate the red and blue zinc ions jointly, while the green zinc ion is coordinated solely by the five-Cys segment.
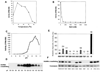
Figure 3. Enzymatic Properties of Recombinant DIM-5
HKMT activity as functions of (A) temperature, (B) salt concentration, (C) pH, and (D) AdoMet crosslinking as a function of pH. The buffers used were 50 mM Na citrate for pH 5.0–6.0, MES for pH 6.0–6.5, HEPES for pH 7.0–7.5, Tris for pH 8.0–8.5, Bicine for pH 9.0, and glycine for pH 9.35–10.7. To rule out the potential inhibitory effect of Na citrate, both Na citrate and Mes are used for pH 6.0. (E) Activities of DIM-5 mutants with conservative point mutations. All mutant proteins were expressed to level similar to that of the wild-type, though some were less soluble, and all were monomeric, suggesting that none of the mutations caused gross aggregation of the protein. Various amounts of mutant enzymes were used, the activities were compared to that of serial dilutions of wild-type enzymes purified in the same way, and the specific activity of mutant proteins relative to wild-type was estimated. The activities shown are averages of at least two measurements. (F) Fluorographic results of an AdoMet crosslinking experiment at pH 8.0 are shown along with results of Coomassie staining to control for the amount of mutant protein tested.
Figure 4. The Cofactor Binding and Active Site
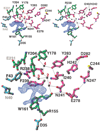
Close-up view of the proposed cofactor binding site and the adjacent active site (top, stereo; bottom, mono). The difference electron density map (blue) is contoured at 5.5σ; the water molecules are numbered 1–4. Dashed lines indicate the hydrogen bonds. The residues are colored as in Figure 1. The water at site 2 is hydrogen bonded to the main chain carbonyl oxygen atom of R238 and to the water molecules at sites 1 and 3, which in turn interacts with the side chain carbonyl oxygen of N241 and the side chain hydroxyl oxygen of Y204, respectively.
Figure 5. Putative Peptide Binding Cleft
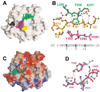
(A) Front view of GRASP surface (Nicholls et al., 1991). The difference electron density map (black) is contoured at 5.5σ. Strand β10 is green, N241, H242, and Y283 are magenta, and C244 is yellow. (B) Superimposition of Drosophila HP1 β strand (light gray) (Jacobs and Khorasanizadeh, 2002; PDBcode 1KNA) and DIM-5 strand β10 (green). Dashed lines indicate the hydrogen bonds between HP1 and H3 peptide (yellow). The DIM-5 residues on the other side of the HP1 peptide are colored magenta. The dimethylated (methyl groups in black) target nitrogen atom occupies water site 2 (see Figure 4). The sequence of histone H3 peptide is shown at the bottom; both K4 and K14 are five residues away from K9. (C) The docked H3 peptide lies in the putative peptide binding cleft. The cleft extends in both directions following turns as indicated. Surface charge distribution is displayed as blue for positive, red for negative, and white for neutral. (D) Superimposition of active site NPPY residues of _Taq_I DNA-adenine amino MTase (gray) (Goedecke et al., 2001; PDB code 1G38) and the proposed DIM-5 active site residues N241, H242, and Y283 (magenta). The Tyr in both cases is hydrogen bonded to a main chain amide nitrogen atom (dashed bonds).
Figure 6. Metal Chelators Inhibit DIM-5 Activity
(A) Analysis of zinc content of DIM-5 with and without EDTA treatment. DIM-5 protein was incubated with 20 mM EDTA for 2 days, at which time HKMT activity was no longer detectable. To remove zinc bound to EDTA, the protein was either dialyzed (Exp1) or subjected to gel filtration chromatography (Exp2) against 20 mM glycine (pH 9.8), 5% glycerol, 0.5 mM DTT, and 1 mM EDTA. (B) Purified DIM-5 protein (1 mg/ml in 20 mM glycine [pH 9.8], 5% glycerol) was incubated with various concentration of 1,10-phenanthroline or EDTA for 18 hr at 4°C. The enzyme was diluted 80-fold and assayed for HKMT activity under standard conditions, except that no DTT was present. (C) Fluorographic results of AdoMet crosslinking in the presence of EDTA.
Similar articles
- Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet.
Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Kwon T, et al. EMBO J. 2003 Jan 15;22(2):292-303. doi: 10.1093/emboj/cdg025. EMBO J. 2003. PMID: 12514135 Free PMC article. - Crystal structure and functional analysis of the histone methyltransferase SET7/9.
Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Wilson JR, et al. Cell. 2002 Oct 4;111(1):105-15. doi: 10.1016/s0092-8674(02)00964-9. Cell. 2002. PMID: 12372304 - Structures of SET domain proteins: protein lysine methyltransferases make their mark.
Yeates TO. Yeates TO. Cell. 2002 Oct 4;111(1):5-7. doi: 10.1016/s0092-8674(02)01010-3. Cell. 2002. PMID: 12372294 Review. - Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes.
Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Ketel CS, et al. Mol Cell Biol. 2005 Aug;25(16):6857-68. doi: 10.1128/MCB.25.16.6857-6868.2005. Mol Cell Biol. 2005. PMID: 16055700 Free PMC article. - Cracking the histone code: one, two, three methyls, you're out!
Dutnall RN. Dutnall RN. Mol Cell. 2003 Jul;12(1):3-4. doi: 10.1016/s1097-2765(03)00282-x. Mol Cell. 2003. PMID: 12887887 Review.
Cited by
- Zinc Metalloproteins in Epigenetics and Their Crosstalk.
Yusuf AP, Abubakar MB, Malami I, Ibrahim KG, Abubakar B, Bello MB, Qusty N, Elazab ST, Imam MU, Alexiou A, Batiha GE. Yusuf AP, et al. Life (Basel). 2021 Feb 26;11(3):186. doi: 10.3390/life11030186. Life (Basel). 2021. PMID: 33652690 Free PMC article. Review. - Histone lysine methylation dynamics: establishment, regulation, and biological impact.
Black JC, Van Rechem C, Whetstine JR. Black JC, et al. Mol Cell. 2012 Nov 30;48(4):491-507. doi: 10.1016/j.molcel.2012.11.006. Mol Cell. 2012. PMID: 23200123 Free PMC article. Review. - Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa.
Adhvaryu KK, Morris SA, Strahl BD, Selker EU. Adhvaryu KK, et al. Eukaryot Cell. 2005 Aug;4(8):1455-64. doi: 10.1128/EC.4.8.1455-1464.2005. Eukaryot Cell. 2005. PMID: 16087750 Free PMC article. - Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112.
Woodcock CB, Yu D, Zhang X, Cheng X. Woodcock CB, et al. Cell Discov. 2019 Sep 10;5:50. doi: 10.1038/s41421-019-0119-5. eCollection 2019. Cell Discov. 2019. PMID: 31632689 Free PMC article. No abstract available. - Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet.
Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Kwon T, et al. EMBO J. 2003 Jan 15;22(2):292-303. doi: 10.1093/emboj/cdg025. EMBO J. 2003. PMID: 12514135 Free PMC article.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. - PMC - PubMed
- Blumenthal RM, Cheng X. A Taq attack displaces bases. Nat. Struct. Biol. 2001;8:101–103. - PubMed
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM061355-03/GM/NIGMS NIH HHS/United States
- GM61355/GM/NIGMS NIH HHS/United States
- R37 GM035690/GM/NIGMS NIH HHS/United States
- R01 GM035690/GM/NIGMS NIH HHS/United States
- GM49245/GM/NIGMS NIH HHS/United States
- GM35690/GM/NIGMS NIH HHS/United States
- R01 GM049245/GM/NIGMS NIH HHS/United States
- R01 GM061355/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases