Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases - PubMed (original) (raw)
. 2002 Dec 10;99 Suppl 4(Suppl 4):16419-26.
doi: 10.1073/pnas.212527999. Epub 2002 Oct 8.
Affiliations
- PMID: 12374855
- PMCID: PMC139903
- DOI: 10.1073/pnas.212527999
Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases
Fabrizio Chiti et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Protein aggregation and the formation of highly insoluble amyloid structures is associated with a range of debilitating human conditions, which include Alzheimer's disease, Parkinson's disease, and the Creutzfeldt-Jakob disease. Muscle acylphosphatase (AcP) has already provided significant insights into mutational changes that modulate amyloid formation. In the present paper, we have used this system to investigate the effects of mutations that modify the charge state of a protein without affecting significantly the hydrophobicity or secondary structural propensities of the polypeptide chain. A highly significant inverse correlation was found to exist between the rates of aggregation of the protein variants under denaturing conditions and their overall net charge. This result indicates that aggregation is generally favored by mutations that bring the net charge of the protein closer to neutrality. In light of this finding, we have analyzed natural mutations associated with familial forms of amyloid diseases that involve alteration of the net charge of the proteins or protein fragments associated with the diseases. Sixteen mutations have been identified for which the mechanism of action that causes the pathological condition is not yet known or fully understood. Remarkably, 14 of these 16 mutations cause the net charge of the corresponding peptide or protein that converts into amyloid deposits to be reduced. This result suggests that charge has been a key parameter in molecular evolution to ensure the avoidance of protein aggregation and identifies reduction of the net charge as an important determinant in at least some forms of protein deposition diseases.
Figures
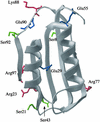
Figure 1
Structure of AcP in its native state. Residues that have been mutated in the present study are labeled and their side chains shown. The various amino acid substitutions are listed in Table 1.
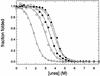
Figure 2
Urea denaturation curves of representative AcP variants in 50 mM acetate buffer, pH 5.5, 28°C. Curves are normalized to the fraction of folded protein and correspond to those of wild-type AcP (filled circles), K88Q (open circles), R23Q (filled squares), E90H (open squares), and S8H (open triangles) mutants. The solid lines through the data represent the best fits of the data points to the equation given by Santoro and Bolen (46). The resulting thermodynamic parameters for all protein variants are listed in Table 1.
Figure 3
(a) Aggregation of six representative AcP variants followed by ThT fluorescence. Aggregation was initiated in each case in 25% TFE/50 mM acetate buffer, pH 5.5, 25°C. Aliquots were withdrawn at regular time intervals for the ThT assay. The AcP variants shown are: wild-type (filled circles), R23Q (open triangles), E29Q (crosses), E29R (open circles), S21R (filled squares), and E90H (diamonds). The solid lines through the data points represent the best fits to single exponential functions. The resulting rate constant values are reported for all variants in Table 1. (b) Aggregation rate versus net charge constructed with the data points of the wild-type protein and the 15 mutants. Changes of net charge on mutation are calculated at pH 5.5 assuming standard pKa values for amino acid residues.
Similar articles
- Rationalization of the effects of mutations on peptide and protein aggregation rates.
Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Chiti F, et al. Nature. 2003 Aug 14;424(6950):805-8. doi: 10.1038/nature01891. Nature. 2003. PMID: 12917692 - Is there a common intracellular bioreactor in which amyloid formation is initiated in neurodegenerative diseases?
Mayer RJ, Landon M, Lowe J, Tipler C, Arnold J, Laszlo L. Mayer RJ, et al. Biochem Soc Trans. 1994 Feb;22(1):151-5. doi: 10.1042/bst0220151. Biochem Soc Trans. 1994. PMID: 8206213 Review. No abstract available. - Relative influence of hydrophobicity and net charge in the aggregation of two homologous proteins.
Calamai M, Taddei N, Stefani M, Ramponi G, Chiti F. Calamai M, et al. Biochemistry. 2003 Dec 30;42(51):15078-83. doi: 10.1021/bi030135s. Biochemistry. 2003. PMID: 14690417 - Dissociation of amyloid fibrils of alpha-synuclein and transthyretin by pressure reveals their reversible nature and the formation of water-excluded cavities.
Foguel D, Suarez MC, Ferrão-Gonzales AD, Porto TC, Palmieri L, Einsiedler CM, Andrade LR, Lashuel HA, Lansbury PT, Kelly JW, Silva JL. Foguel D, et al. Proc Natl Acad Sci U S A. 2003 Aug 19;100(17):9831-6. doi: 10.1073/pnas.1734009100. Epub 2003 Aug 4. Proc Natl Acad Sci U S A. 2003. PMID: 12900507 Free PMC article. - Curcumin: A small molecule with big functionality against amyloid aggregation in neurodegenerative diseases and type 2 diabetes.
Radbakhsh S, Barreto GE, Bland AR, Sahebkar A. Radbakhsh S, et al. Biofactors. 2021 Jul;47(4):570-586. doi: 10.1002/biof.1735. Epub 2021 Apr 24. Biofactors. 2021. PMID: 33893674 Review.
Cited by
- Misfolding and amyloid aggregation of apomyoglobin.
Iannuzzi C, Maritato R, Irace G, Sirangelo I. Iannuzzi C, et al. Int J Mol Sci. 2013 Jul 9;14(7):14287-300. doi: 10.3390/ijms140714287. Int J Mol Sci. 2013. PMID: 23839096 Free PMC article. Review. - An in silico model of the ubiquitin-proteasome system that incorporates normal homeostasis and age-related decline.
Proctor CJ, Tsirigotis M, Gray DA. Proctor CJ, et al. BMC Syst Biol. 2007 Mar 21;1:17. doi: 10.1186/1752-0509-1-17. BMC Syst Biol. 2007. PMID: 17408507 Free PMC article. - Dissimilar roles of the four conserved acidic residues in the thermal stability of poly(A)-specific ribonuclease.
He GJ, Liu WF, Yan YB. He GJ, et al. Int J Mol Sci. 2011;12(5):2901-16. doi: 10.3390/ijms12052901. Epub 2011 May 3. Int J Mol Sci. 2011. PMID: 21686157 Free PMC article. Review. - PrP charge structure encodes interdomain interactions.
Martínez J, Sánchez R, Castellanos M, Makarava N, Aguzzi A, Baskakov IV, Gasset M. Martínez J, et al. Sci Rep. 2015 Sep 1;5:13623. doi: 10.1038/srep13623. Sci Rep. 2015. PMID: 26323476 Free PMC article. - Lack of Dependence of the Sizes of the Mesoscopic Protein Clusters on Electrostatics.
Vorontsova MA, Chan HY, Lubchenko V, Vekilov PG. Vorontsova MA, et al. Biophys J. 2015 Nov 3;109(9):1959-68. doi: 10.1016/j.bpj.2015.09.025. Biophys J. 2015. PMID: 26536272 Free PMC article.
References
- Kelly J W. Curr Opin Struct Biol. 1996;6:11–17. - PubMed
- Plante-Bordeneuve V, Said G. Curr Opin Neurol. 2000;13:569–573. - PubMed
- Uemichi T, Liepnieks J J, Alexander F, Benson M D. Q J Med. 1996;89:745–750. - PubMed
- Benson M D, Liepnieks J, Uemichi T, Wheeler G, Correa R. Nat Genet. 1993;3:252–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases