N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation - PubMed (original) (raw)
N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation
Paul S Knoepfler et al. Genes Dev. 2002.
Abstract
To address the role of N-myc in neurogenesis and in nervous system tumors, it was conditionally disrupted in neuronal progenitor cells (NPCs) with a nestin-Cre transgene. Null mice display ataxia, behavioral abnormalities, and tremors that correlate with a twofold decrease in brain mass that disproportionately affects the cerebellum (sixfold reduced in mass) and the cerebral cortex, both of which show signs of disorganization. In control mice at E12.5, we observe a domain of high N-Myc protein expression in the rapidly proliferating cerebellar primordium. Targeted deletion of N-myc results in severely compromised proliferation as shown by a striking decrease in S phase and mitotic cells as well as in cells expressing the Myc target gene cyclin D2, whereas apoptosis is unaffected. Null progenitor cells also have comparatively high levels of the cdk inhibitors p27(Kip1) and p18(Ink4c), whereas p15(Ink4b), p21(Cip1), and p19(Ink4d) levels are unaffected. Many null progenitors also exhibit altered nuclear morphology and size. In addition, loss of N-myc disrupts neuronal differentiation as evidenced by ectopic staining of the neuron specific marker betaTUBIII in the cerebrum. Furthermore, in progenitor cell cultures derived from null embryonic brain, we observe a dramatic increase in neuronal differentiation compared with controls. Thus, N-myc is essential for normal neurogenesis, regulating NPC proliferation, differentiation, and nuclear size. Its effects on proliferation and differentiation appear due, at least in part, to down-regulation of a specific subset of cyclin-dependent kinase inhibitors.
Figures
Figure 1
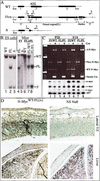
Nestin-Cre(+).N-_myc_FL/FL mice are nervous system-specific N-myc nulls. (A) Wild-type (WT), Floxed (Flox), and deleted (Δ) N-myc alleles. The thick line above the Flox allele is the Southern probe. (Eco*) The introduced _Eco_RI site just 5′ of the loxP site. loxP sites are represented as arrowheads. Numbers and arrows above the alleles represent PCR primers. (B) Southern blotting of _Eco_RI-digested DNA from ES cells (left) and the mice generated from the ES cells (right). (Lane 1) Wild-type ES cells. FL/WT ES cells (lane 2) were used to generate chimeric mice that were subsequently used to produce FL/WT F1 mice (lane 4). FL/WT mice were then interbred, producing F2 mice: WT (lane 6), FL/WT (lane 7) and FL/FL (lane 8). Cre was introduced into FL/WT ES cells to produce true Δ/WT ES cells (lane 3). (C) RNA and DNA samples were isolated simultaneously from whole brains of P20 pups (lanes 1–4) and E19 embryos (lanes 5–8). Genomic PCR was conducted on DNA samples (top), FL/WT with (lanes 2,6) and without nestin-Cre (lanes 1,5) as well as FL/FL with (lanes 4,8) and without nestin-Cre (lanes 3,7). RT–PCR for N-myc, L-myc, and c-myc as well as Rpl7 (ribosomal protein gene used as control) was conducted on total RNA samples from the same brains (bottom). (D) N-Myc staining of E12.5 embryos.
Figure 2
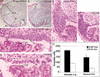
N-myc deficiency results in reduced brain size. (A) Total body mass of animals from P8 to P32. (B) Total brain mass of animals from 8 to 16 wk of age. (C,D) External views of E12.5 control and null littermate embryos, respectively. Asterisks indicate forebrain area. (E,F) H&E stain of control and null E12.5 head regions, respectively.
Figure 3
Lack of N-myc severely disrupts cerebellar development. (A–F) H&E staining of P21 cerebella. (A,C) Control cerebella. (B,D) Null cerebella. (E,F) Folia of control and null cerebella. (G) Absolute cerebellar mass in milligrams from more than four animals of each genotype. (H) Cerebellar mass relative to total brain mass. In D, X is a reference spot to refer to the equivalent position in Figure 4B. (I) Diagram of murine cerebellar development [based on data in Altman and Bayer (1997)]. (Top) Rostral; (bottom) caudal. (NE) neuroepithelium; (EGL) external germinal layer; (ML) molecular layer; (PCL) Purkinje cell layer; (GCL) granular cell layer. Magnification: A_–_D, 5×; E,F, 40×.
Figure 4
The N-myc NS null E17.5 cerebellum primordium is small and exhibits germinal cell deficiency. (A,B) H&E-stained control and null cerebella primordia, respectively. (C,D) Control and null neuroepithelium. (E,F) Control and null rhombic lip. (G) Cell counts, the mean of three sections, from the rhombic lip and EGL. RL, rhombic lip; ChP, choroid plexus; EGL, external germinal layer; ne, neuroepithelium. Magnification: A,B, 5×; C,D, 100×; E,F, 40×.
Figure 5
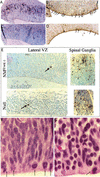
The N-myc NS null E12–E13 cerebellum primordium exhibits NPC deficiency, loss of cyclin D2 expression, and ectopic CDKI expression. Sections of cerebella primordia were stained as indicated. The arrow in A identifies the domain at the apex of the cerebellum primordium that lacks significant N-myc, cyclinD2, and BrdU staining.
Figure 6
Analysis of mitosis, S phase, and apoptosis in the E13 lateral VZ. (A,C) BrdU staining of a control and null. (B,D) Anti-phosphoH3 staining of a control and null. (E) TUNEL staining. (F,G) The dorsal portion of the lateral VZ in a control and null embryo, respectively. Arrows point to mitotic figures (A,F,G) or apoptic cells (E). Magnification: E, 40×; F,G, 100×.
Figure 7
N-Myc inhibits neuronal differentiation. (A,B) H&E (left) and βTubulinIII (right) staining of the control and null E17.5 cerebral cortex in the lateral ventricle region. Cells positively stained for β-tubulinIII are brown. (C,D) Anti-βTubulinIII immunofluorescence of control and null E12.5 neurosphere cultures stimulated to differentiate for 5 d. (E,F) Anti-GFAP immunofluorescence of control and null E12.5 neurosphere cultures stimulated to differentiate for 5 d. (G) Nuclear size in microns of the same neurosphere cultures as above. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; MZ, marginal zone.
Similar articles
- The cyclin-dependent kinase inhibitors p19(Ink4d) and p27(Kip1) are coexpressed in select retinal cells and act cooperatively to control cell cycle exit.
Cunningham JJ, Levine EM, Zindy F, Goloubeva O, Roussel MF, Smeyne RJ. Cunningham JJ, et al. Mol Cell Neurosci. 2002 Mar;19(3):359-74. doi: 10.1006/mcne.2001.1090. Mol Cell Neurosci. 2002. PMID: 11906209 - Consistent inactivation of p19(Arf) but not p15(Ink4b) in murine myeloid cells transformed in vivo by deregulated c-Myc.
Haviernik P, Schmidt M, Hu X, Wolff L. Haviernik P, et al. Oncogene. 2003 Mar 20;22(11):1600-10. doi: 10.1038/sj.onc.1206268. Oncogene. 2003. PMID: 12642863 - The v-myc oncogene.
Lee CM, Reddy EP. Lee CM, et al. Oncogene. 1999 May 13;18(19):2997-3003. doi: 10.1038/sj.onc.1202786. Oncogene. 1999. PMID: 10378695 Review. - Use of transgenic mice to study myc family gene function in normal mammalian development and in cancer.
Morgenbesser SD, DePinho RA. Morgenbesser SD, et al. Semin Cancer Biol. 1994 Feb;5(1):21-36. Semin Cancer Biol. 1994. PMID: 8186385 Review.
Cited by
- Beyond the Warburg Effect: N-Myc Contributes to Metabolic Reprogramming in Cancer Cells.
Yoshida GJ. Yoshida GJ. Front Oncol. 2020 May 27;10:791. doi: 10.3389/fonc.2020.00791. eCollection 2020. Front Oncol. 2020. PMID: 32547946 Free PMC article. Review. - Induction of _MYCN_-amplified neuroblastoma differentiation through NMYC suppression using PPAR-γ antagonist.
Nakao-Ise Y, Narita T, Miyamoto S, Watanabe M, Tanaka T, Sowa Y, Iizumi Y, Masuda M, Fujii G, Hirai Y, Nakao T, Takakura H, Mutoh M. Nakao-Ise Y, et al. J Clin Biochem Nutr. 2023 Nov;73(3):191-197. doi: 10.3164/jcbn.23-28. Epub 2023 Jun 28. J Clin Biochem Nutr. 2023. PMID: 37970556 Free PMC article. - Constitutive gray hair in mice induced by melanocyte-specific deletion of c-Myc.
Pshenichnaya I, Schouwey K, Armaro M, Larue L, Knoepfler PS, Eisenman RN, Trumpp A, Delmas V, Beermann F. Pshenichnaya I, et al. Pigment Cell Melanoma Res. 2012 May;25(3):312-25. doi: 10.1111/j.1755-148X.2012.00998.x. Pigment Cell Melanoma Res. 2012. PMID: 22420299 Free PMC article. - c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development.
Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. Wey A, et al. Cerebellum. 2010 Dec;9(4):537-47. doi: 10.1007/s12311-010-0190-9. Cerebellum. 2010. PMID: 20658325 Free PMC article. - Formation of a structurally-stable conformation by the intrinsically disordered MYC:TRRAP complex.
Feris EJ, Hinds JW, Cole MD. Feris EJ, et al. PLoS One. 2019 Dec 2;14(12):e0225784. doi: 10.1371/journal.pone.0225784. eCollection 2019. PLoS One. 2019. PMID: 31790487 Free PMC article.
References
- Altman J, Bayer S. Development of the cerebellar system: In relation to its evolution, structure, and functions. Boca Raton, FL: CRC Press; 1997. pp. 82–137.
- Badiali M, Pession A, Basso G, Andreini L, Rigobello L, Galassi E, Giangaspero F. N-myc and c-myc oncogenes amplification in medulloblastomas. Evidence of particularly aggressive behavior of a tumor with c-myc amplification. Tumori. 1991;77:118–121. - PubMed
- Bernard O, Drago J, Sheng H. L-myc and N-myc influence lineage determination in the central nervous system. Neuron. 1992;9:1217–1224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous