Nucleic acid-independent retrovirus assembly can be driven by dimerization - PubMed (original) (raw)
Nucleic acid-independent retrovirus assembly can be driven by dimerization
Marc C Johnson et al. J Virol. 2002 Nov.
Abstract
The Gag protein of retroviruses alone can polymerize into regular virus-like particles (VLPs) both in vitro and in vivo. In most circumstances the capsid (CA) and nucleocapsid (NC) domains of Gag as well as some form of nucleic acid are required for this process. The mechanism by which NC-nucleic acid interaction promotes assembly has remained obscure. We show here that while deletion of the NC domain of Rous sarcoma virus Gag abolishes formation and budding of VLPs at the plasma membranes of baculovirus-infected insect cells, replacement of NC with a dimer-forming leucine zipper domain restores budding of spherical particles morphologically similar to wild-type VLPs. The positioning of the dimerization domain appears to be critical for proper assembly, as the insertion of a 5-amino-acid flexible linker upstream of the zipper domain leads to budding of tubular rather than spherical particles. Similar tubular particles are formed when the same linker is inserted upstream of NC. The tubes are morphologically distinct from tubes formed when the p10 domain upstream of CA is deleted. The fact that a foreign dimerization domain can functionally mimic NC suggests that the role of nucleic acid in retroviral assembly is not to serve as a scaffold but rather to promote the formation of Gag dimers, which are critical intermediates in the polymerization of the Gag shell.
Figures
FIG. 1.
Schematic representation of expressed proteins. MA-NC corresponds to the RSV Gag protein lacking its PR domain at the C terminus.
FIG. 2.
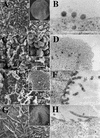
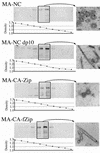
EM images of budding structures. Cells were infected with baculovirus vectors expressing either MA-NC (A and B), MA-CA (C and D), MA-CA-Zip (E and F), or MA-NC-fZip (G and H). Panels A, C, E, and G are SEM images (insets are low-magnification images), and panels B, D, F, and H are thin-section TEM images. Bars, 500 nm.
FIG. 3.
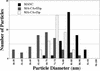
Size distribution of MA-NC, MA-CA-Zip, and MA-CA-fZip. Diameters of spherical (MA-NC and MA-CA-Zip) or tubular (MA-CA-fZip) particles were measured in TEM thin sections.
FIG. 4.
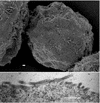
EM images of budding structures of MA-CA-fNC. (Top) SEM; (Bottom) TEM. Bar, 500 nm.
FIG. 5.
EM images of p10 budding structures. Cells were infected with baculovirus vectors expressing either MA-NC dp10 (A and B) or MA-NC dp10 + 25 AA (C and D). (A and C) SEM images; (B and D) thin-section TEM images. Bars, 500 nm.
FIG. 6.
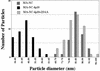
Size distribution of MA-NC, MA-NC dp10, and MA-NC dp10 + 25 AA particles. Diameters of spherical (MA-NC and MA-NC dp10 + 25 AA) or tubular (MA-NC dp10) particles were measured in TEM thin sections.
FIG. 7.
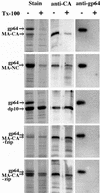
Detergent stability of particles. Particles were pelleted after incubation with or without Triton X-100 (Tx-100) as described in Materials and Methods. Pellets were dissolved in SDS, and the proteins were separated by SDS-PAGE and then either stained with Coomassie blue, probed with an antibody against RSV CA, or probed with an antibody against baculovirus gp64.
FIG. 8.
Density of particles. Pelleted particles from each expression construct were resuspended and then centrifuged to equilibrium in 10-to-60% (wt/wt) sucrose gradients. A portion of each fraction was separated by SDS-PAGE and probed with an antibody against RSV CA. The density of each fraction was determined by refractometry. Fractions from the boxed lanes were pooled, diluted to reduce the sucrose concentration, and then centrifuged again to collect the particles. The pellets were then viewed by thin-section TEM and are shown on the right.
Similar articles
- Role of the Rous sarcoma virus p10 domain in shape determination of gag virus-like particles assembled in vitro and within Escherichia coli.
Joshi SM, Vogt VM. Joshi SM, et al. J Virol. 2000 Nov;74(21):10260-8. doi: 10.1128/jvi.74.21.10260-10268.2000. J Virol. 2000. PMID: 11024160 Free PMC article. - Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro.
Ma YM, Vogt VM. Ma YM, et al. J Virol. 2004 Jan;78(1):52-60. doi: 10.1128/jvi.78.1.52-60.2004. J Virol. 2004. PMID: 14671087 Free PMC article. - Characterization of Rous sarcoma virus Gag particles assembled in vitro.
Yu F, Joshi SM, Ma YM, Kingston RL, Simon MN, Vogt VM. Yu F, et al. J Virol. 2001 Mar;75(6):2753-64. doi: 10.1128/JVI.75.6.2753-2764.2001. J Virol. 2001. PMID: 11222698 Free PMC article. - Nucleic acid chaperone activity of retroviral Gag proteins.
Rein A. Rein A. RNA Biol. 2010 Nov-Dec;7(6):700-5. doi: 10.4161/rna.7.6.13685. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045546 Free PMC article. Review. - Properties and functions of the nucleocapsid protein in virus assembly.
Muriaux D, Darlix JL. Muriaux D, et al. RNA Biol. 2010 Nov-Dec;7(6):744-53. doi: 10.4161/rna.7.6.14065. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21157181 Free PMC article. Review.
Cited by
- Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein.
Nadaraia-Hoke S, Bann DV, Lochmann TL, Gudleski-O'Regan N, Parent LJ. Nadaraia-Hoke S, et al. J Virol. 2013 Mar;87(6):3609-15. doi: 10.1128/JVI.03059-12. Epub 2013 Jan 16. J Virol. 2013. PMID: 23325682 Free PMC article. - Dynamic Association between HIV-1 Gag and Membrane Domains.
Hogue IB, Llewellyn GN, Ono A. Hogue IB, et al. Mol Biol Int. 2012;2012:979765. doi: 10.1155/2012/979765. Epub 2012 Jul 5. Mol Biol Int. 2012. PMID: 22830021 Free PMC article. - A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance.
Porterfield JZ, Zlotnick A. Porterfield JZ, et al. Virology. 2010 Nov 25;407(2):281-8. doi: 10.1016/j.virol.2010.08.015. Epub 2010 Sep 17. Virology. 2010. PMID: 20850162 Free PMC article. - Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy.
Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. Bharat TA, et al. Nature. 2012 Jul 19;487(7407):385-9. doi: 10.1038/nature11169. Nature. 2012. PMID: 22722831 - APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein.
Wang SM, Wang CT. Wang SM, et al. Virology. 2009 May 25;388(1):112-20. doi: 10.1016/j.virol.2009.03.010. Epub 2009 Apr 5. Virology. 2009. PMID: 19345973 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources