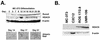Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter - PubMed (original) (raw)
Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter
Jennifer J Westendorf et al. Mol Cell Biol. 2002 Nov.
Abstract
Runx2 (Cbfa1, AML-3) is multifunctional transcription factor that is essential for osteoblast development. Runx2 binds specific DNA sequences and interacts with transcriptional coactivators and corepressors to either activate or repress transcription of tissue-specific genes. In this study, the p21(CIP/WAF1) promoter was identified as a repressible target of Runx2. A carboxy-terminal repression domain distinct from the well-characterized TLE/Groucho-binding domain contributed to Runx2-mediated p21 repression. This carboxy-terminal domain was sufficient to repress a heterologous GAL reporter. The repressive activity of this domain was sensitive to the histone deacetylase inhibitor trichostatin A but not to trapoxin B. HDAC6, which is insensitive to trapoxin B, specifically interacted with the carboxy terminus of Runx2. The HDAC6 interaction domain of Runx2 was mapped to a region overlapping the nuclear matrix-targeting signal. The Runx2 carboxy terminus was necessary for recruitment of HDAC6 from the cytoplasm to chromatin. HDAC6 also colocalized and coimmunoprecipitated with the nuclear matrix-associated protein Runx2 in osteoblasts. Finally, we show that HDAC6 is expressed in differentiating osteoblasts and that the Runx2 carboxy terminus is necessary for maximal repression of the p21 promoter in preosteoblasts. These data identify Runx2 as the first transcription factor to interact with HDAC6 and suggest that HDAC6 may bind to Runx2 in differentiating osteoblasts to regulate tissue-specific gene expression.
Figures
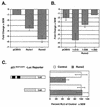
FIG. 1.
Runx2 represses the p21 promoter. (A) Runx1 and Runx2 repress the p21 promoter. NIH-3T3 cells were cotransfected with p21-Luc, pCMV5-SEAP, and either pCMV5, pCMV5-Runx1, or pCMV5-Runx2 (MRIPV isoform). (B) Two regions in the Runx2 carboxy terminus are necessary for complete repression of the p21 promoter. NIH-3T3 cells were cotransfected with p21-Luc, pCMV5-SEAP, and either pCMV5-M2 or the indicated pCMV5-M2-Runx2 expression construct. (C) Runx binding sites in the p21 promoter are required for repression by Runx2. ROS17/2.8 cells were transfected with the indicated reporter plasmids and either pCMV5 (Control) or pCMV5-Runx2 (MRIPV). Runx binding sites are indicated by black boxes. The basal activities of the reporters differed significantly; thus the relative light units (RLU) in the Runx2-expressing lysates are shown as percentages of that in the control. Values represent the mean of triplicate samples; error bars indicate standard errors of the means (SEM).
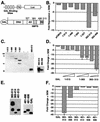
FIG. 2.
The Runx2 carboxy terminus contains several portable repression domains. (A) Schematics of the heterologous GAL-Luc reporter and GAL-Runx2 (1-513) fusion proteins used in these experiments. The reporter contains a minimal herpes virus TK promoter between four GAL4 binding sites and the Luc reporter gene. Amino acid residue numbers are above the Runx2 diagram. The relative locations of the activation domain (AD), nuclear matrix targeting signal (NMTS), and the TLE binding repression domain (RD) are depicted. (B) The TLE binding domain is not required for GAL-Runx2-mediated repression of a heterologous reporter. NIH-3T3 cells were cotransfected with GAL-TK-Luc, pRL-TK, and 700 ng of the indicated pCMV5-M2 or pCMV5-M2-Runx2 expression constructs. Firefly luciferase activities were corrected for transfection efficiency with renilla luciferase values. The change in repression (_n_-fold) was calculated relative to that in cells transfected with pCMV5. (C) The relative expression levels of GAL-Runx2 fusion proteins were determined by immunoblot analysis with anti-GAL-DNA binding domain monoclonal antibody. (D) Runx2 contains multiple regions flanking the activation domain that contribute to repression. In these experiments, pCMV5-M2-Runx2 plasmids were added in various quantities (100, 300, or 700 ng). Values were corrected to SEAP activity, and the change in repression (_n_-fold) was calculated relative to that in cells transfected with pCMV5-GAL. (E) The relative expression levels of GAL-Runx2-C-terminal fusion proteins were determined by immunoblot analysis as described for panel C. (F) The nuclear matrix targeting signal is also a portable transcriptional repressor. These experiments were performed as described above for panel D.
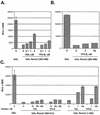
FIG. 3.
Repression by the Runx2 carboxy terminus is sensitive to the HDAC inhibitor TSA. (A) The complete Runx2 carboxy terminus containing the TLE binding region and a second repression domain is partially sensitive to TSA. NIH-3T3 cells were transfected with GAL-TK-Luc, pCMV-SEAP, and 700 ng of pCMV5-Runx2 (383-513) in the presence or absence of the indicated concentrations of TSA. Luciferase values were corrected for transfection efficiency with SEAP values. The change in repression (_n_-fold) was calculated relative to that in cells expressing nonfused GAL (i.e., those transfected with pCMV-M2). (B) Runx2 (383-498) is completely sensitive to TSA. NIH-3T3 cells were transfected with the indicated pCMV5-M2-Runx2 constructs as described above and cultured in the presence (black bars) or absence (white bars) of 300 nM TSA.
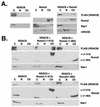
FIG. 4.
HDAC6 interacts with the Runx2 carboxy terminus. (A) The Runx2 carboxy terminus interacts with HDAC6 but not with HDAC2, HDAC4, or HDAC5. COS7 cells were cotransfected with pCMV5-M2-Runx2 (383-513) in the presence or absence of the indicated pCMV5-FLAG-HDAC construct. FLAG immunoprecipitates and a fraction (3%) of the whole-cell lysates (WCL) were resolved by SDS-PAGE. To detect Runx2 and the HDACs, membranes were immunoblotted with GAL and FLAG monoclonal antibodies, respectively. (B) HDAC6 specifically interacts with the Runx2 carboxy terminus. COS7 cells were cotransfected with pCMV5-FLAG-HDAC6 and the indicated pCMV5-M2-Runx2 construct. (C) Runx2 residues (383-414) are sufficient to interact with HDAC6. (D) Runx2 residues (461-513) do not interact with HDAC6. In panels C and D, COS cells were transfected as described for panel B. Proteins were detected as described above for panel A. Many of the GAL-Runx2 expression plasmids produce doublet proteins. The upper band represents the protein of the correct molecular weight.
FIG. 5.
Runx2 repression domains are differentially sensitive to TSA and TPX-B. (A and B) The second Runx2-carboxy-terminal repression domain is sensitive to TSA but not to TPX-B. NIH-3T3 cells were cotransfected with pCMV5-GAL-TK-Luc, pCMV5-SEAP, and pCMV5-M2 or pCMV-M2-Runx2 (383-498). Transfected cells were cultured in the presence or absence of the indicated concentrations of TSA or TPX-B. (C) The complete Runx2 carboxy terminus (383-513) is partially insensitive to both TSA and TPX-B, but the remainder of Runx2 (1-383) is sensitive to both HDAC inhibitors. NIH-3T3 cells were transfected and cultured as described above. RLU, relative light units.
FIG. 6.
Runx2 and HDAC6 coassociate on chromatin. (A) Runx2 recruits HDAC6 to chromatin. COS7 cells were transfected with pCMV5-FLAG-HDAC6, pCMV5-Runx2, or both plasmids. Cells were lysed as described in Materials and Methods to collect cytoplasmic (C), nuclear (N), or chromatin-containing (Ch) fractions. Proteins were resolved by SDS-PAGE and detected by immunoblotting with FLAG or Runx2 antibodies. Blots were also incubated with antibodies specific for Rac1 and mSinA, the cytoplasmic and nuclear proteins, respectively, to monitor the fractionation procedure. (B) The Runx2 carboxy terminus is required for HDAC6 association with chromatin. COS7 cells were transfected with pCMV5-FLAG-HDAC6 (top panel) or pCMV5-FLAG-HDAC5 (bottom panel) in the absence or presence of pCMV5-Runx2 (1-513) (full-length) or pCMV5-Runx2 (1-383). Proteins were extracted and detected as described for panel A.
FIG. 7.
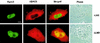
Runx2 and HDAC6 coassociate in the nucleus. HeLa cells were cotransfected with pCMV5-Runx2 and pCMV5-FLAG-HDAC6 and cultured in the presence or absence of the nuclear export inhibitor LMB prior to immunofluorescence detection. Bar, 10 μm.
FIG. 8.
HDAC6 is expressed in differentiating osteoblasts. (A) MC3T3 cells were differentiated in ascorbic acid and β-glycerolphosphate for the indicated number of days. Total cell lysates (100 μg) were resolved by SDS-PAGE. Runx2, HDAC6, and β-actin were detected by immunoblotting with protein-specific antisera. Calcium matrix production in differentiating osteoblasts was determined in parallel cultures of MC-3T3 cells by staining with Alizarin Red on days 14, 17, and 21 of differentiation. Calcium nodules are indicative of localized matrix production and differentiation. (B) HDAC6 is expressed in mature osteoblast lineage cell lines. Twenty micrograms of lysates from MC3T3-E1 (preosteoblast), C2C12 (osteoblast/myocyte precursor), ROS17/2.8, and UMR-106 (osteosarcomas with a mature osteoblast phenotype) cells were resolved by SDS-PAGE. HDAC6 and β-actin were detected by immunoblotting.
FIG. 9.
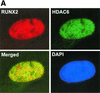
Runx2 and HDAC6 complexes are present in osteoblast lineage cells. (A) Runx2 and HDAC6 colocalize in ROS17/2.8 cell nuclei. ROS17/2.8 cells were grown on gelatin-coated coverslips, fixed, lysed, and incubated with Runx2- and HDAC6-specific antisera and DAPI. Proteins were detected by in situ immunofluorescence microscopy. Digital images were analyzed and deconvolved with the Metamorph software program. (B) HDAC6 and Runx2 coimmunoprecipitate in vivo. ROS17/2.8 lysates were incubated with HDAC6 (Cell Signaling Technology) or Runx2 (S-19; Santa Cruz) antibodies or no antibody (Control). Protein A Sepharose beads were added to all reactions. Proteins bound to the beads were resolved by SDS-PAGE and immunoblotted with HDAC6 and Runx2 antisera.
FIG. 9.
Runx2 and HDAC6 complexes are present in osteoblast lineage cells. (A) Runx2 and HDAC6 colocalize in ROS17/2.8 cell nuclei. ROS17/2.8 cells were grown on gelatin-coated coverslips, fixed, lysed, and incubated with Runx2- and HDAC6-specific antisera and DAPI. Proteins were detected by in situ immunofluorescence microscopy. Digital images were analyzed and deconvolved with the Metamorph software program. (B) HDAC6 and Runx2 coimmunoprecipitate in vivo. ROS17/2.8 lysates were incubated with HDAC6 (Cell Signaling Technology) or Runx2 (S-19; Santa Cruz) antibodies or no antibody (Control). Protein A Sepharose beads were added to all reactions. Proteins bound to the beads were resolved by SDS-PAGE and immunoblotted with HDAC6 and Runx2 antisera.
FIG. 10.
The second repression domain in the Runx2 carboxy terminus is required for maximal repression of the p21 promoter in preosteoblasts. MC3T3 cells were transfected with p21-Luc, pCMV5-SEAP, and pCMV5-M2 or the indicated pCMV-M2-Runx2 expression plasmid. Values represent the means of triplicate samples.
Similar articles
- Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner.
Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Jensen ED, et al. J Bone Miner Res. 2008 Mar;23(3):361-72. doi: 10.1359/jbmr.071104. J Bone Miner Res. 2008. PMID: 17997710 Free PMC article. - Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation.
Schroeder TM, Kahler RA, Li X, Westendorf JJ. Schroeder TM, et al. J Biol Chem. 2004 Oct 1;279(40):41998-2007. doi: 10.1074/jbc.M403702200. Epub 2004 Aug 2. J Biol Chem. 2004. PMID: 15292260 - Modulation of the cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa Cells.
Liu PY, Chan JY, Lin HC, Wang SL, Liu ST, Ho CL, Chang LC, Huang SM. Liu PY, et al. Mol Cancer Res. 2008 Jul;6(7):1204-14. doi: 10.1158/1541-7786.MCR-08-0123. Mol Cancer Res. 2008. PMID: 18644983 - Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites.
Sowa Y, Orita T, Hiranabe-Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Sowa Y, et al. Ann N Y Acad Sci. 1999;886:195-9. doi: 10.1111/j.1749-6632.1999.tb09415.x. Ann N Y Acad Sci. 1999. PMID: 10667218 Review. - Histone deacetylase inhibitors: signalling towards p21cip1/waf1.
Ocker M, Schneider-Stock R. Ocker M, et al. Int J Biochem Cell Biol. 2007;39(7-8):1367-74. doi: 10.1016/j.biocel.2007.03.001. Epub 2007 Mar 7. Int J Biochem Cell Biol. 2007. PMID: 17412634 Review.
Cited by
- RUNX2 Phosphorylation by Tyrosine Kinase ABL Promotes Breast Cancer Invasion.
He F, Matsumoto Y, Asano Y, Yamamura Y, Katsuyama T, La Rose J, Tomonobu N, Komalasari NLGY, Sakaguchi M, Rottapel R, Wada J. He F, et al. Front Oncol. 2021 May 31;11:665273. doi: 10.3389/fonc.2021.665273. eCollection 2021. Front Oncol. 2021. PMID: 34136397 Free PMC article. - Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner.
Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Jensen ED, et al. J Bone Miner Res. 2008 Mar;23(3):361-72. doi: 10.1359/jbmr.071104. J Bone Miner Res. 2008. PMID: 17997710 Free PMC article. - Post-translational Regulation of Runx2 in Bone and Cartilage.
Jonason JH, Xiao G, Zhang M, Xing L, Chen D. Jonason JH, et al. J Dent Res. 2009 Aug;88(8):693-703. doi: 10.1177/0022034509341629. J Dent Res. 2009. PMID: 19734454 Free PMC article. Review. - The influence of the morphology of titania and hydroxyapatite on the proliferation and osteogenic differentiation of human mesenchymal stem cells.
Kuvyrkou YU, Brezhneva N, Skorb EV, Ulasevich SA. Kuvyrkou YU, et al. RSC Adv. 2021 Jan 20;11(7):3843-3853. doi: 10.1039/d0ra08271f. eCollection 2021 Jan 19. RSC Adv. 2021. PMID: 35424371 Free PMC article. - Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications.
Krajnović M, Kožik B, Božović A, Jovanović-Ćupić S. Krajnović M, et al. Cells. 2023 Sep 19;12(18):2303. doi: 10.3390/cells12182303. Cells. 2023. PMID: 37759525 Free PMC article. Review.
References
- Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. - PMC - PubMed
- Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 66:1-10. - PubMed
- Bertos, N. R., A. H. Wang, and X. J. Yang. 2001. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79:243-252. - PubMed
- Blyth, K., A. Terry, N. Mackay, F. Vaillant, M. Bell, E. R. Cameron, J. C. Neil, and M. Stewart. 2001. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene 20:295-302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources