Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis - PubMed (original) (raw)
Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis
Julie D Bouchard et al. J Bacteriol. 2002 Nov.
Abstract
During the production of fermented dairy products, virulent bacteriophages infecting Lactococcus lactis can delay or stop the milk acidification process. A solution to this biological problem consists of introducing natural phage barriers into the strains used by the dairy industry. One such hurdle is called abortive infection (Abi) and causes premature cell death with no or little phage progeny. Here, we describe the isolation and characterization of a novel Abi mechanism encoded by plasmid pED1 from L. lactis. The system is composed of two constitutively cotranscribed genes encoding putative proteins of 127 and 213 amino acids, named AbiTi and AbiTii, respectively. Site-directed mutagenesis indicated that a hydrophobic region at the C-terminal extremity of AbiTi is essential to the antiphage phenotype. The AbiT system is effective against phages of the 936 and P335 species (efficiency of plaquing between 10(-5) and 10(-7)) and causes a 20-fold reduction in the efficiency to form centers of infection as well as a 10- to 12-fold reduction in the burst size. Its efficacy could be improved by raising the plasmid copy number, but changing the intrinsic ratio of AbiTi and AbiTii did not greatly affect the antiphage activity. The monitoring of the intracellular phage infection process by DNA replication, gene expression, and electron microscopy as well as the study of phage mutants by genome mapping indicated that AbiT is likely to act at a later stage of the phage lytic cycle.
Figures
FIG. 1.
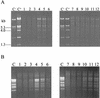
Identification and expression of the genetic determinants of AbiT encoded on pED1. (A) Restriction map and pertinent subclones of pED1 with their respective Abi phenotype. The smallest Abi+ region obtained is delimited by a gray box. (B) DNA sequence analysis of the 2,711-bp _Hae_II fragment and inactivation of the ORFs. The putative promoter is indicated by a thin arrow, and predicted genes are indicated by large arrows. Regions of homology with lactococcal plasmids are shown in gray; the hatched box is a region of homology with E. faecalis and S. pneumoniae transposons. X, frameshift. (C) Hybridization of RNA isolated from AbiT-containing L. lactis strains with a labeled DNA fragment of abiTi, abiTii, and orf98-orf109. Lane 1, MG1363(pED100); lane 2, MG1363(pED107). The size of the transcripts was evaluated with a 0.24- to 9.5-kb RNA ladder (Invitrogen).
FIG. 2.
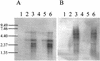
DNA replication of phages p2 (A) and ul36 (B) in AbiT− (left) and AbiT+ (right) cells. (A) C−, unheated p2 DNA; C+, p2 DNA heated (10 min at 65°C). Lanes 1 to 6 and 7 to 12, total DNA from infected MG1363(pMIG3) and MG1363(pED109) cells, respectively, at t = 0, 10, 20, 30, 40, and 50 min, heated for 10 min at 65°C. (B) C, ul36 DNA. Lanes 1 to 6 and 7 to 12, total DNA from infected SMQ481(pMIG3) and SMQ481(pED109) cells, respectively, at t = 0, 20, 40, 60, 80, and 100 min. All samples were digested with _Eco_RV.
FIG. 3.
Hybridization of RNA from p2-infected AbiT− (left) and AbiT+ (right) cells with the orf35 homologue (A) and the mcp gene (B). Lanes 1, 2, and 3, MG1363(pMIG3) at t = 0, 10, and 25 min, respectively; lanes 4, 5, and 6, MG1363(pED109) at t = 0, 10, and 25 min.
FIG. 4.
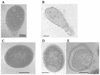
Monitoring of the lytic cycle by electron microscopy. (A and B) L. lactis AbiT− cells infected with phage sk1 at 25 and 45 min, respectively. (C) L. lactis AbiT+ cells infected with phage sk1 at 25 min. (D and E) L. lactis AbiT+ cells infected at 45 min.
FIG. 5.
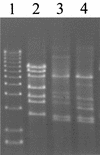
_Eco_RV restriction profile of AbiT-insensitive mutants of phage ul36. Lane 1, 1-kb DNA ladder (1.6 to 12.2 kb; Invitrogen); lane 2, ul36; lane 3, ul36T1; lane 4, ul36T2 DNA.
Similar articles
- Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis.
Emond E, Holler BJ, Boucher I, Vandenbergh PA, Vedamuthu ER, Kondo JK, Moineau S. Emond E, et al. Appl Environ Microbiol. 1997 Apr;63(4):1274-83. doi: 10.1128/aem.63.4.1274-1283.1997. Appl Environ Microbiol. 1997. PMID: 9097424 Free PMC article. - Microbiological and molecular impacts of AbiK on the lytic cycle of Lactococcus lactis phages of the 936 and P335 species.
Boucher I, Émond É, Dion É, Montpetit D, Moineau S. Boucher I, et al. Microbiology (Reading). 2000 Feb;146 ( Pt 2):445-453. doi: 10.1099/00221287-146-2-445. Microbiology (Reading). 2000. PMID: 10708383 - Molecular characterization of a new abortive infection system (AbiU) from Lactococcus lactis LL51-1.
Dai G, Su P, Allison GE, Geller BL, Zhu P, Kim WS, Dunn NW. Dai G, et al. Appl Environ Microbiol. 2001 Nov;67(11):5225-32. doi: 10.1128/AEM.67.11.5225-5232.2001. Appl Environ Microbiol. 2001. PMID: 11679349 Free PMC article. - Phage abortive infection in lactococci: variations on a theme.
Chopin MC, Chopin A, Bidnenko E. Chopin MC, et al. Curr Opin Microbiol. 2005 Aug;8(4):473-9. doi: 10.1016/j.mib.2005.06.006. Curr Opin Microbiol. 2005. PMID: 15979388 Review. - Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome.
Bolotin A, Mauger S, Malarme K, Ehrlich SD, Sorokin A. Bolotin A, et al. Antonie Van Leeuwenhoek. 1999 Jul-Nov;76(1-4):27-76. Antonie Van Leeuwenhoek. 1999. PMID: 10532372 Review.
Cited by
- Tail assembly interference is a common strategy in bacterial antiviral defenses.
He L, Miguel-Romero L, Patkowski JB, Alqurainy N, Rocha EPC, Costa TRD, Fillol-Salom A, Penadés JR. He L, et al. Nat Commun. 2024 Aug 30;15(1):7539. doi: 10.1038/s41467-024-51915-4. Nat Commun. 2024. PMID: 39215040 Free PMC article. - Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages.
Labrie SJ, Moineau S. Labrie SJ, et al. J Bacteriol. 2007 Feb;189(4):1482-7. doi: 10.1128/JB.01111-06. Epub 2006 Oct 13. J Bacteriol. 2007. PMID: 17041060 Free PMC article. - Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages.
Samson JE, Moineau S. Samson JE, et al. Appl Environ Microbiol. 2010 Oct;76(20):6843-52. doi: 10.1128/AEM.00796-10. Epub 2010 Aug 27. Appl Environ Microbiol. 2010. PMID: 20802084 Free PMC article. - Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages.
Samson JE, Bélanger M, Moineau S. Samson JE, et al. J Bacteriol. 2013 Sep;195(17):3947-56. doi: 10.1128/JB.00296-13. J Bacteriol. 2013. PMID: 23813728 Free PMC article. - AbiV, a novel antiphage abortive infection mechanism on the chromosome of Lactococcus lactis subsp. cremoris MG1363.
Haaber J, Moineau S, Fortier LC, Hammer K. Haaber J, et al. Appl Environ Microbiol. 2008 Nov;74(21):6528-37. doi: 10.1128/AEM.00780-08. Epub 2008 Sep 5. Appl Environ Microbiol. 2008. PMID: 18776030 Free PMC article.
References
- Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226.
- Behnke, D., and H. Malke. 1978. Bacteriophage interference in Streptococcus pyogenes. I. Characterization of prophage-host systems interfering with the virulent phage A25. Virology 85:118-128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources