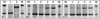RNA stable isotope probing, a novel means of linking microbial community function to phylogeny - PubMed (original) (raw)
RNA stable isotope probing, a novel means of linking microbial community function to phylogeny
Mike Manefield et al. Appl Environ Microbiol. 2002 Nov.
Abstract
Identifying microorganisms responsible for recognized environmental processes remains a great challenge in contemporary microbial ecology. Only in the last few years have methodological innovations provided access to the relationship between the function of a microbial community and the phylogeny of the organisms accountable for it. In this study stable-isotope-labeled [13C]phenol was fed into a phenol-degrading community from an aerobic industrial bioreactor, and the 13C-labeled RNA produced was used to identify the bacteria responsible for the process. Stable-isotope-labeled RNA was analyzed by equilibrium density centrifugation in concert with reverse transcription-PCR and denaturing gradient gel electrophoresis. In contradiction with findings from conventional methodologies, this unique approach revealed that phenol degradation in the microbial community under investigation is dominated by a member of the Thauera genus. Our results suggest that this organism is important for the function of this bioreactor.
Figures
FIG. 1.
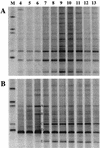
RNA-SIP analysis of a defined, mixed bacterial culture. (A) GC-clamped 16S rRNA (V3 region) RT-PCR products from pure cultures of a phenol-degrading isolate (P. putida; lane 1) and a nondegrading isolate (P. chlororaphis; lane 2) on a denaturing gradient gel. (B) RT-PCR products from fractions 5, 7, 9, and 11 (lanes 1 to 4, respectively) of an equilibrium density gradient loaded with RNA extracted at 24 h from a mixed P. putida, P. chlororaphis culture grown on phenol-13C6. RNA from both species was detected in fractions 9 and 11 (associated by buoyant density with unlabeled RNA). The 16S rRNA sequence of P. putida alone was detectable in fractions 5 and 7 (13C-labeled RNA), demonstrating specific access to 13C according to metabolic activity. (C) RT-PCR products from fractions 5, 7, 9, and 11 (lanes 1 to 4, respectively) of a gradient loaded with RNA extracted after 72 h from the mixed culture. RNA from both species was detectable in all four fractions, indicative of 13C cross-feeding between species. These results are representative of two separate experiments. Lanes labeled M were loaded with an unrelated DGGE marker.
FIG. 2.
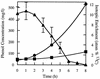
Degradation of phenol-13C6 and increase in 13C atom percent of RNA and DNA in the microbial community from an industrial bioreactor. Bioreactor samples were pulsed with 500 μg of phenol-13C6/ml and were assessed hourly for phenol concentration (triangles). The phenol concentration remained constant over the initial 2 h but then dropped rapidly over the following 5 h. A reduced phenol degradation rate preceded the complete removal of the pulse by 8 h. Samples were assayed for phenol in triplicate. Error bars represent standard errors. These data are representative of three separate experiments. Total community DNA and RNA samples were extracted at 0, 1, 2, 4, and 8 h and were subjected to IRMS analysis. The 13C atom percent of RNA (•) increased more than 10-fold as the labeled phenol pulse was utilized, while that of DNA (▪) increased just over 2-fold. Nucleic acid samples were analyzed in triplicate from a single experiment. Error bars representing standard deviation were too small to be visible.
FIG. 3.
RNA-SIP profiles relating community function to specific microbial community components. DGGE profile of community RNA present in fractions 4 to 13 from equilibrium density gradients loaded with RNA extracted 1 (A) and 8 h (B) after the phenol-13C6 pulse was administered. Lane numbers correspond to fraction numbers. Five of the most dominant bands are labeled A to E (B). After 1 h the profile suggests that RNA from all detectable species have the same buoyant density prior to phenol degradation. After 8 h the profile suggests that RNA from particular community members (bands A, B, and D) have increased in buoyant density subsequent to phenol degradation. Lanes labeled M were loaded with an unrelated DGGE marker.
FIG. 4.
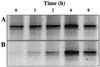
Increasing buoyant density of the dominant band in 16S rRNA community profiles throughout the duration of the phenol-13C6 pulse. (A) The intensity of the dominant DGGE band remains constant in fraction 10 over time. (B) The intensity of the dominant DGGE band increases in fraction 6 over time, confirming an increase in the buoyant density of RNA from this species as phenol-13C6 is consumed.
Similar articles
- Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function.
Manefield M, Whiteley AS, Ostle N, Ineson P, Bailey MJ. Manefield M, et al. Rapid Commun Mass Spectrom. 2002;16(23):2179-83. doi: 10.1002/rcm.782. Rapid Commun Mass Spectrom. 2002. PMID: 12442292 - Microorganisms involved in anaerobic phenol degradation in the treatment of synthetic coke-oven wastewater detected by RNA stable-isotope probing.
Sueoka K, Satoh H, Onuki M, Mino T. Sueoka K, et al. FEMS Microbiol Lett. 2009 Feb;291(2):169-74. doi: 10.1111/j.1574-6968.2008.01448.x. FEMS Microbiol Lett. 2009. PMID: 19146573 - Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community.
DeRito CM, Pumphrey GM, Madsen EL. DeRito CM, et al. Appl Environ Microbiol. 2005 Dec;71(12):7858-65. doi: 10.1128/AEM.71.12.7858-7865.2005. Appl Environ Microbiol. 2005. PMID: 16332760 Free PMC article. - Stable isotope probing - linking microbial identity to function.
Dumont MG, Murrell JC. Dumont MG, et al. Nat Rev Microbiol. 2005 Jun;3(6):499-504. doi: 10.1038/nrmicro1162. Nat Rev Microbiol. 2005. PMID: 15886694 Review. - Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms.
Radajewski S, McDonald IR, Murrell JC. Radajewski S, et al. Curr Opin Biotechnol. 2003 Jun;14(3):296-302. doi: 10.1016/s0958-1669(03)00064-8. Curr Opin Biotechnol. 2003. PMID: 12849783 Review.
Cited by
- A culture-independent approach to unravel uncultured bacteria and functional genes in a complex microbial community.
Wang Y, Chen Y, Zhou Q, Huang S, Ning K, Xu J, Kalin RM, Rolfe S, Huang WE. Wang Y, et al. PLoS One. 2012;7(10):e47530. doi: 10.1371/journal.pone.0047530. Epub 2012 Oct 17. PLoS One. 2012. PMID: 23082176 Free PMC article. - Unearthing the Ecology of Soil Microorganisms Using a High Resolution DNA-SIP Approach to Explore Cellulose and Xylose Metabolism in Soil.
Pepe-Ranney C, Campbell AN, Koechli CN, Berthrong S, Buckley DH. Pepe-Ranney C, et al. Front Microbiol. 2016 May 12;7:703. doi: 10.3389/fmicb.2016.00703. eCollection 2016. Front Microbiol. 2016. PMID: 27242725 Free PMC article. - Flow-through stable isotope probing (Flow-SIP) minimizes cross-feeding in complex microbial communities.
Mooshammer M, Kitzinger K, Schintlmeister A, Ahmerkamp S, Nielsen JL, Nielsen PH, Wagner M. Mooshammer M, et al. ISME J. 2021 Jan;15(1):348-353. doi: 10.1038/s41396-020-00761-5. Epub 2020 Sep 2. ISME J. 2021. PMID: 32879458 Free PMC article. - mRNA-based parallel detection of active methanotroph populations by use of a diagnostic microarray.
Bodrossy L, Stralis-Pavese N, Konrad-Köszler M, Weilharter A, Reichenauer TG, Schöfer D, Sessitsch A. Bodrossy L, et al. Appl Environ Microbiol. 2006 Feb;72(2):1672-6. doi: 10.1128/AEM.72.2.1672-1676.2006. Appl Environ Microbiol. 2006. PMID: 16461725 Free PMC article. - Nitrate removal performance and diversity of active denitrifying bacteria in denitrification reactors using poly(L-lactic acid) with enhanced chemical hydrolyzability.
Yamada T, Tsuji H, Daimon H. Yamada T, et al. Environ Sci Pollut Res Int. 2019 Dec;26(36):36236-36247. doi: 10.1007/s11356-019-06722-6. Epub 2019 Nov 11. Environ Sci Pollut Res Int. 2019. PMID: 31713134
References
- Anders, H.-J., A. Kaetzke, P. Kampfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying Pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Intl. J. Syst. Bacteriol. 45:327-333. - PubMed
- Birnie, G. D. 1978. Centrifugal methods for characterising macromolecules and their interactions, p. 95-125. In G. D. Birnie and D. Rickwood (ed.), Centrifugal separations in molecular and cell biology. Butterworths, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases