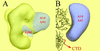The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits - PubMed (original) (raw)
The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits
Gerald Peyroche et al. Proc Natl Acad Sci U S A. 2002.
Abstract
A43, an essential subunit of yeast RNA polymerase I (pol I), interacts with Rrn3, a class I general transcription factor required for rDNA transcription. The pol I-Rrn3 complex is the only form of enzyme competent for promoter-dependent transcription initiation. In this paper, using biochemical and genetic approaches, we demonstrate that the A43 polypeptide forms a stable heterodimer with the A14 pol I subunit and interacts with the common ABC23 subunit, the yeast counterpart of the omega subunit of bacterial RNA polymerase. We show by immunoelectronic microscopy that A43, ABC23, and A14 colocalize in the three-dimensional structure of the pol I, and we demonstrate that the presence of A43 is required for the stabilization of both A14 and ABC23 within the pol I. Because the N-terminal half of A43 is clearly related to the pol II Rpb7 subunit, we propose that the A43-A14 pair is likely the pol I counterpart of the Rpb7-Rpb4 heterodimer, although A14 distinguishes from Rpb4 by specific sequence and structure features. This hypothesis, combined with our structural data, suggests a new localization of Rpb7-Rpb4 subunits in the three-dimensional structure of yeast pol II.
Figures
Fig 1.
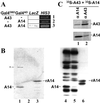
Subunit A43 interacts with subunit A14. (A) Two-hybrid. Y190 yeast strain was transformed by two plasmids, one driving the expression of a Gal4DBD fusion protein, the other driving the expression of a Gal4AD fusion protein (as indicated). Activation of the LacZ and HIS3 reporter genes was monitored by blue color in the presence of X-gal and by growth in the presence of 50 mM 3-aminotriazol, respectively. (B) Far Western blot. Ten micrograms each of pol I (lanes 1 and 4) and pol IΔ (lanes 2 and 5) and 1 μg of purified recombinant, tagged A14 subunit (lanes 3 and 6) were subjected to SDS/PAGE and transferred onto poly(vinylidene difluoride) membranes. The membrane was incubated with 35S-labeled A43 (lanes 1–3), and radiolabeled proteins were visualized by autoradiography. The asterisk indicates a nonspecific signal that does not correspond to a pol I subunit. Subunits of pol I, pol IΔ, and recombinant A14 subunit were identified by Western blot using anti-pol I antibodies (lanes 4–6, respectively). (C) Coimmunoprecipitation. 35S-A43 and 35S-A14 polypeptides were preincubated and subjected to immunoprecipitation by antibodies raised against subunit A14 or A43 (αA14 and αA43). Immunoprecipitated proteins were separated by SDS/PAGE and identified by autoradiography.
Fig 2.
Genetic interactions between RPA43 and RPA14 genes. (A) Positions and nature of the mutated residues in the rpa43 thermosensitive mutant are indicated. In A43-18, the C terminus of the protein is replaced by a short divergent amino acid sequence (thick line). Growth phenotype at different temperatures conferred by these different mutant alleles is summarized on the right. (B) Multicopy suppression. The rpa43-6, rpa43-18, and rpa43-4 mutant strains were transformed with a centromeric plasmid carrying the wild-type RPA43 gene (lanes 1, 4, and 7), with the empty pFL44 multicopy plasmid as a control (lanes 2, 5, and 8), or with the pFL44 vector harboring the RPA14 gene (lanes 3, 6, and 9). Growth of two dilutions of transformants was monitored at restrictive temperature.
Fig 3.
Interaction domains of subunit A43 with subunit A14. (A) Interaction of A14 subunit with hemagglutinin-tagged A43 polypeptides progressively truncated at the N terminus (A43-ΔN) or at the C terminus (A43-ΔC) (the number of deleted residues is indicated). Each truncated form was synthesized in vitro in the presence of [35S]methionine (no protein was obtained for the ΔN80 construction), and their interaction with 35S-A14 was monitored by immunoprecipitation with anti-hemagglutinin antibodies. Immunoprecipitated proteins were separated by SDS/PAGE and visualized by autoradiography. (B) The ORFs encoding the truncated versions of subunit A43 were cloned in fusion with Gal4DBD in pGBT9 vector. Their ability to interact with A14 fused to Gal4AD was monitored in Y190 strain. Activation of the LacZ and HIS3 reporter genes was monitored by staining in the presence of X-gal and by analysis of growth in the presence of 50 mM 3-aminotriazol, respectively. (C) Schematic representation of the interaction domains of A43 with A14. The conserved domain of A43 (residues 42–168), present in putative orthologues of this subunit in Schizosaccharomyces pombe, Candida albicans, and higher eukaryotes (see ref. 22), is indicated. The 87 C-terminal residues of A43 are not conserved through evolution (data not shown). These residues are not essential for cell viability (see Fig. 2) but contain an interaction domain with A14.
Fig 4.
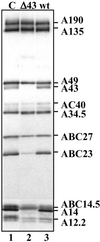
Subunit composition of pol I partially purified from the _rpa43_-Δ strain. The subunit composition of pol I partially purified from WT (lane 3) and _rpa43_-Δ strains (lane 2) was analyzed by Western blot using antibodies raised against yeast pol I (lane 1, purified pol I).
Fig 5.
The A43 subunit interacts with the ABC23 subunit. Presence of A43 subunit was checked by Western blot using anti-A43 antibodies in crude cell extracts prepared from an E. coli strain coexpressing A43 and His-6-tagged ABC23 (lanes 2, 4, and 6), and from a control strain expressing only A43 subunit (lanes 1, 3, and 5). The asterisk indicates a proteolyzed form of A43. Lanes 3 and 4, extracts were loaded onto a Nickel column, bound proteins were eluted by competition with 200 mM imidazole, and the presence of A43 subunit in the elution fractions was checked by Western blot analysis using anti-A43. Lanes 5 and 6, Ponceau red staining on the membrane after transfer of the proteins of the elution fractions.
Fig 6.
Colocalization of subunits A43, ABC23, and A14 within the pol I. (A) Structure of the yeast pol I as determined by cryoelectron microscopy at 25 Å resolution (gray shading) (31). Blue tags represent the positions of subunits A14 and A43, mapped by immunolabeling (19, 31), and of Rpb6, inferred from the docking of the atomic structure of pol IIΔ 4/7 (43) into the pol I envelope. These results show that subunits A14 and A43 form a stalk near Rpb6. (B) Close-up view of the interaction interface of A14 and A43 with the core enzyme. The blue volume represents the additional density due to A14 and A43 present in pol I. The aligned atomic structure of pol II shows that the C-terminal repeats of Rpb1 (CTD) start next to the interaction site of the two pol I–specific subunits.
Fig 7.
The A43/A14 subunits belong to the rpoE/rpoF family. (A) Sequence alignment of S. cerevisiae A43, C25, and Rpb7 subunits, and Methanococcus jannaschii rpoE subunit, whose secondary structures are reported (see text). Sequence identities are indicated on a black background; similarities are gray shaded (white letters, hydrophobic amino acids or amino acids that can substitute them in some circumstances). RpoE residues with <10% of their surface accessible to solvent in the rpoE/rpoF structure are indicated with stars (open stars for residues participating in the rpoE hydrophobic core and filled stars for residues involved in contacts with rpoF). (B) Sequence alignment of the conserved motifs of the S. cerevisiae A14 subunit (133 residues) and of its putative homologue from C. albicans (177 residues). Predicted secondary structures are shown up to the sequences and labeled according to their putative correspondence to the rpoF structure. (C) Ribbon representation of the rpoE–rpoF dimer (41). Circled regions represent regions predicted to be different in the A43–A14 subunit. (D) A possible model for the domain structure of the A43–A14 subunit (see text).
Similar articles
- Structural and functional homology between the RNAP(I) subunits A14/A43 and the archaeal RNAP subunits E/F.
Meka H, Daoust G, Arnvig KB, Werner F, Brick P, Onesti S. Meka H, et al. Nucleic Acids Res. 2003 Aug 1;31(15):4391-400. doi: 10.1093/nar/gkg652. Nucleic Acids Res. 2003. PMID: 12888498 Free PMC article. - The fission yeast protein Ker1p is an ortholog of RNA polymerase I subunit A14 in Saccharomyces cerevisiae and is required for stable association of Rrn3p and RPA21 in RNA polymerase I.
Imazawa Y, Hisatake K, Mitsuzawa H, Matsumoto M, Tsukui T, Nakagawa K, Nakadai T, Shimada M, Ishihama A, Nogi Y. Imazawa Y, et al. J Biol Chem. 2005 Mar 25;280(12):11467-74. doi: 10.1074/jbc.M411150200. Epub 2005 Jan 12. J Biol Chem. 2005. PMID: 15647272 - The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3.
Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. Peyroche G, et al. EMBO J. 2000 Oct 16;19(20):5473-82. doi: 10.1093/emboj/19.20.5473. EMBO J. 2000. PMID: 11032814 Free PMC article. - Rpb4 and Rpb7: subunits of RNA polymerase II and beyond.
Choder M. Choder M. Trends Biochem Sci. 2004 Dec;29(12):674-81. doi: 10.1016/j.tibs.2004.10.007. Trends Biochem Sci. 2004. PMID: 15544954 Review. - Synthesis of the ribosomal RNA precursor in human cells: mechanisms, factors and regulation.
Daiß JL, Griesenbeck J, Tschochner H, Engel C. Daiß JL, et al. Biol Chem. 2023 Jul 17;404(11-12):1003-1023. doi: 10.1515/hsz-2023-0214. Print 2023 Oct 26. Biol Chem. 2023. PMID: 37454246 Review.
Cited by
- Ancient origin, functional conservation and fast evolution of DNA-dependent RNA polymerase III.
Proshkina GM, Shematorova EK, Proshkin SA, Zaros C, Thuriaux P, Shpakovski GV. Proshkina GM, et al. Nucleic Acids Res. 2006 Jul 28;34(13):3615-24. doi: 10.1093/nar/gkl421. Print 2006. Nucleic Acids Res. 2006. PMID: 16877568 Free PMC article. - RNA polymerase I-specific subunit CAST/hPAF49 has a role in the activation of transcription by upstream binding factor.
Panov KI, Panova TB, Gadal O, Nishiyama K, Saito T, Russell J, Zomerdijk JC. Panov KI, et al. Mol Cell Biol. 2006 Jul;26(14):5436-48. doi: 10.1128/MCB.00230-06. Mol Cell Biol. 2006. PMID: 16809778 Free PMC article. - Architecture of initiation-competent 12-subunit RNA polymerase II.
Armache KJ, Kettenberger H, Cramer P. Armache KJ, et al. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):6964-8. doi: 10.1073/pnas.1030608100. Epub 2003 May 13. Proc Natl Acad Sci U S A. 2003. PMID: 12746495 Free PMC article. - Site specific phosphorylation of yeast RNA polymerase I.
Gerber J, Reiter A, Steinbauer R, Jakob S, Kuhn CD, Cramer P, Griesenbeck J, Milkereit P, Tschochner H. Gerber J, et al. Nucleic Acids Res. 2008 Feb;36(3):793-802. doi: 10.1093/nar/gkm1093. Epub 2007 Dec 15. Nucleic Acids Res. 2008. PMID: 18084032 Free PMC article. - Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III.
Tan Q, Prysak MH, Woychik NA. Tan Q, et al. Mol Cell Biol. 2003 May;23(9):3329-38. doi: 10.1128/MCB.23.9.3329-3338.2003. Mol Cell Biol. 2003. PMID: 12697831 Free PMC article.
References
- Udem S. A. & Warner, J. R. (1972) J. Mol. Biol. 65, 227-242. - PubMed
- Dequard-Chablat M., Riva, M., Carles, C. & Sentenac, A. (1991) J. Biol. Chem. 266, 15300-15307. - PubMed
- Carles C., Treich, I., Bouet, F., Riva, M. & Sentenac, A. (1991) J. Biol. Chem. 266, 24092-24096. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases