Natural selection of tumor variants in the generation of "tumor escape" phenotypes - PubMed (original) (raw)
Review
Natural selection of tumor variants in the generation of "tumor escape" phenotypes
Hung T Khong et al. Nat Immunol. 2002 Nov.
Abstract
The idea that tumors must "escape" from immune recognition contains the implicit assumption that tumors can be destroyed by immune responses either spontaneously or as the result of immunotherapeutic intervention. Simply put, there is no need for tumor escape without immunological pressure. Here, we review evidence supporting the immune escape hypothesis and critically explore the mechanisms that may allow such escape to occur. We discuss the idea that the central engine for generating immunoresistant tumor cell variants is the genomic instability and dysregulation that is characteristic of the transformed genome. "Natural selection" of heterogeneous tumor cells results in the survival and proliferation of variants that happen to possess genetic and epigenetic traits that facilitate their growth and immune evasion. Tumor escape variants are likely to emerge after treatment with increasingly effective immunotherapies.
Figures
Figure 1. Activation versus suppression during tumor progression
Immune activation during early tumor progression may be triggered by the expression of neoantigens. In addition, the progressive growth of tumors may be associated with the invasion and destruction of surrounding normal tissues. Other factors that may cause immune activation include generation of heat shock proteins (Hsps), which result from cellular stress, and ROS such as OH− and H2O2. Conversely, immune activation and function may be hampered by a lack of appropriate costimulation, the presence of immunosuppressive cytokines (for example, VEGF, IL-10 and TGF-β), and the impact of immunoregulatory cells such as CD4+CD25+ T cells and NKT cells. As tumors grow, these events occur in the presence of massive apoptosis and necrosis of tumor cells, infiltrating immune cells and surrounding stromal cells. The fate of tumors may be dictated by the net effect of immune activation and immune inhibition.
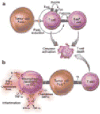
Figure 2. Natural selection of tumor variants in the generation of “tumor escape” phenotypes
(a) Genomic instability gives rise to genetic diversity in tumors. Natural selection of tumor variants occurs by differential propagation of tumor subclones in their microenvironment. (b) The same concept also applies to tumor growth after effective immunotherapy.
Figure 3. Molecular mechanisms responsible for HLA class I deficiency
Tumor antigens are processed in the proteosome and generate peptides that are transported by TAP to the endoplasmic reticulum (ER); there they bind to certain HLA class I heavy chain in association with β2-microglobulin. The peptide-HLA complexes are then transported through the Golgi to the cell surface. Several different defects are associated with tumor “escape” from immune recognition: (a) defects in components of the antigen-processing machinery (such as LMP-2 and LMP-7) in the proteosome result in HLA class I down-regulation; (b) defects in peptide transporters TAP-1 or TAP-2 cause HLA class I down-regulation; and (c) LOH on chromosome 6 causes HLA class I haplotype loss. Defects in the transcriptional regulation of the HLA class I gene result in HLA class I locus loss. Point mutations or gene deletions involving the HLA class I heavy chain result in HLA class I allelic loss. (d) β2-microglobulin (β2M) mutation or deletion results in total loss of HLA class I.
Figure 4. Alternative models for the induction of FasL-mediated T cell death after encounter with tumor cells
(a) AICD of T cells after recognition of tumor cells. Tumor recognition leads to activation of T cells and up-regulation of Fas and FasL on T cell surface, which results in the T cell killing of itself (“suicide”) and of other T cells (“fratricide”). (b) Proposed tumor “counterattack” model. Tumor cells express functional FasL and kill infiltrating Fas-expressing T cells via Fas-FasL binding, which leads to tumor escape. However, ligation of Fas expressed on innate immune cells such as neutrophils, macrophages and immature DCs by FasL expressed on tumor cells may also lead to release of multiple proinflammatory cytokines and chemokines, setting the stage for tumor rejection.
Similar articles
- Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance.
Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Marincola FM, et al. Adv Immunol. 2000;74:181-273. doi: 10.1016/s0065-2776(08)60911-6. Adv Immunol. 2000. PMID: 10605607 Review. No abstract available. - The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape.
Algarra I, García-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. Algarra I, et al. Cancer Immunol Immunother. 2004 Oct;53(10):904-10. doi: 10.1007/s00262-004-0517-9. Epub 2004 Apr 7. Cancer Immunol Immunother. 2004. PMID: 15069585 Free PMC article. Review. - [Immunoediting eventually proven in humans. Genetics to assist immunotherapy].
David Fumet J, Ghiringhelli F. David Fumet J, et al. Med Sci (Paris). 2019 Aug-Sep;35(8-9):629-631. doi: 10.1051/medsci/2019125. Epub 2019 Sep 18. Med Sci (Paris). 2019. PMID: 31532373 French. No abstract available. - MHC class I antigens, immune surveillance, and tumor immune escape.
Garcia-Lora A, Algarra I, Garrido F. Garcia-Lora A, et al. J Cell Physiol. 2003 Jun;195(3):346-55. doi: 10.1002/jcp.10290. J Cell Physiol. 2003. PMID: 12704644 Review. - Commentary: Immune escape versus tumor tolerance: how do tumors evade immune surveillance?
Salih HR, Nüssler V. Salih HR, et al. Eur J Med Res. 2001 Aug 27;6(8):323-32. Eur J Med Res. 2001. PMID: 11549514 Review.
Cited by
- A comprehensive pan-cancer analysis of the expression characteristics, prognostic value, and immune characteristics of TOP1MT.
Fei L, Lu Z, Xu Y, Hou G. Fei L, et al. Front Genet. 2022 Aug 10;13:920897. doi: 10.3389/fgene.2022.920897. eCollection 2022. Front Genet. 2022. PMID: 36035140 Free PMC article. - Cellular immunotherapy for carcinoma using genetically modified EGFR-specific T lymphocytes.
Zhou X, Li J, Wang Z, Chen Z, Qiu J, Zhang Y, Wang W, Ma Y, Huang N, Cui K, Li J, Wei YQ. Zhou X, et al. Neoplasia. 2013 May;15(5):544-53. doi: 10.1593/neo.13168. Neoplasia. 2013. PMID: 23633926 Free PMC article. - Gene therapy for advanced melanoma: selective targeting and therapeutic nucleic acids.
Viola JR, Rafael DF, Wagner E, Besch R, Ogris M. Viola JR, et al. J Drug Deliv. 2013;2013:897348. doi: 10.1155/2013/897348. Epub 2013 Mar 25. J Drug Deliv. 2013. PMID: 23634303 Free PMC article. - The Potential of Tumor Debulking to Support Molecular Targeted Therapies.
Oppel F, Görner M, Sudhoff H. Oppel F, et al. Front Oncol. 2020 Jun 18;10:801. doi: 10.3389/fonc.2020.00801. eCollection 2020. Front Oncol. 2020. PMID: 32626653 Free PMC article. - Effect of miR-513a-5p on etoposide-stimulating B7-H1 expression in retinoblastoma cells.
Wu L, Chen Z, Zhang J, Xing Y. Wu L, et al. J Huazhong Univ Sci Technolog Med Sci. 2012 Aug;32(4):601-606. doi: 10.1007/s11596-012-1004-8. Epub 2012 Aug 11. J Huazhong Univ Sci Technolog Med Sci. 2012. PMID: 22886978
References
- Girardi M, et al. Regulation of cutaneous malignancy by γδT cells. Science. 2001;294:605–609. - PubMed
- Shankaran V, et al. IFN γ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources