The fission yeast pfh1(+) gene encodes an essential 5' to 3' DNA helicase required for the completion of S-phase - PubMed (original) (raw)
The fission yeast pfh1(+) gene encodes an essential 5' to 3' DNA helicase required for the completion of S-phase
Hiroyuki Tanaka et al. Nucleic Acids Res. 2002.
Abstract
The Cdc24 protein plays an essential role in chromosomal DNA replication in the fission yeast Schizosaccharomyces pombe, most likely via its direct interaction with Dna2, a conserved endonuclease-helicase protein required for Okazaki fragment processing. To gain insights into Cdc24 function, we isolated cold-sensitive chromosomal suppressors of the temperature-sensitive cdc24-M38 allele. One of the complementation groups of such suppressors defined a novel gene, pfh1(+), encoding an 805 amino acid nuclear protein highly homologous to the Saccharomyces cerevisiae Pif1p and Rrm3p DNA helicase family proteins. The purified Pfh1 protein displayed single-stranded DNA-dependent ATPase activity as well as 5' to 3' DNA helicase activity in vitro. Reverse genetic analysis in S.pombe showed that helicase activity was essential for the function of the Pfh1 protein in vivo. Schizosaccharomyces pombe cells carrying the cold-sensitive pfh1-R20 allele underwent cell cycle arrest in late S/G2-phase of the cell cycle when shifted to the restrictive temperature. This arrest was dependent upon the presence of a functional late S/G2 DNA damage checkpoint, suggesting that Pfh1 is required for the completion of DNA replication. Furthermore, at their permissive temperature pfh1-R20 cells were highly sensitive to the DNA-alkylating agent methyl methanesulphonate, implying a further role for Pfh1 in the repair of DNA damage.
Figures
Figure 1
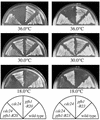
Suppression of the temperature sensitivity of the cdc24-M38 by the pfh1-R20 and pfh1-R23 mutations. The indicated cells were streaked on YE plates and incubated for 3 days at 30°C, 3 days at 36°C or 5 days at 18°C, respectively.
Figure 2
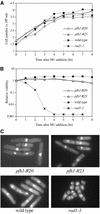
Properties of pfh1-R20 cells. (A) Morphology of pfh1-R20 cells. pfh1-R20 and pfh1 + cells were grown to mid-log phase in minimal medium at 34°C and shifted down to 18°C. Cells were collected at 24 h after temperature shift, fixed with 70% ethanol and stained with DAPI. (B) Flow cytometric analysis of pfh1-R20 cells. Exponentially growing cells at 34°C were shifted down to 18°C as indicated in (A). Samples were taken at indicated times after temperature shift and analyzed by flow cytometry. (C) DAPI staining of pfh1-R20 rad1-1 cells. pfh1-R20 rad1-1 cells from microcolonies formed at 34°C were inoculated on YE plates and incubated at 18°C for 24 h. The cells were then fixed with 70% ethanol and stained with DAPI. Arrows indicate cells that underwent aberrant mitosis. (D) Pulsed-field gel electrophoresis of the chromosomes. Indicated strains growing exponentially at 34°C in minimal medium supplemented with leucine were shifted down to 18°C and incubated for 16 h. Samples were prepared from these cells and exponentially growing cells at 34°C. HU indicates wild-type cells treated with 12 mM HU at 34°C for 4 h. Chromosomes were separated with pulsed-field gel electrophoresis. (E) pfh1-R20 cells are sensitive to MMS and HU. Approximately 104, 103, 102 and 10 cells (from left to right) of exponentially growing pfh1-R20 and wild-type cells in YEL (32) at 34°C were spotted in YE plates or YE plates containing MMS (0.0025, 0.005%), HU (6, 8 mM) or irradiated by UV (150, 200 J/m2). Plates were incubated at 34°C for 2 days (without drug) or 3 days (with drug or UV irradiated).
Figure 3
HU sensitivity of pfh1 mutant cells. (A) Growth curve after HU addition. HU (12 mM, final concentration) was added to asynchronous culture of the indicated strains at 34°C in minimal medium supplemented with leucine. Cell aliquots were taken every hour. (B) pfh1-R20 and pfh1-R23 cells are sensitive to HU. The viability was assayed by colony formation and expressed as relative viability compared with the viability at 0 h. (C) Cell morphology of indicated strains. Cells cultured in the presence of 12 mM HU for 6 h were fixed with 70% ethanol and stained with DAPI.
Figure 4
Isolation of the pfh1 + gene, structure of Pfh1 protein and positions of mutations. (A) Isolation of pfh1 +. Cold-sensitive pfh1-R20 cells carrying indicated plasmids were streaked on an MM (30) plate and incubated at the indicated temperatures. (B) pREP81-pfh1 K338R was unable to suppress pfh1-R20 cells. pfh1-R20 cells carrying the indicated plasmids were incubated as in (A). (C) Structure of Pfh1 protein is shown by an open bar. Seven conserved helicase motifs are indicated by shaded boxes. The part of predicted amino acid sequence of S.pombe pfh1 +, containing motifs III and IV, is shown in single letter code and aligned with Pif1p and Rrm3p of S.cerevisiae. Identical amino acids are boxed. The mutations occurred in pfh1-R20 and pfh1-R23 alleles are indicated.
Figure 5
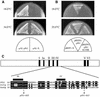
Analysis of the pfh1 + gene and Δ_pfh1_ cells. (A) Restriction map of the pfh1 + gene. The ORF, pfh1 + cDNA and the positions of two MCB sequences are shown. The gene contains one intron. The Eco_RV DNA fragment was replaced with a ura4 + gene cassette for generating Δ_pfh1 cells. (B) pfh1 + is an essential gene. Tetrads generated from pfh1 + /pfh1::ura4 + diploid cells were dissected on YE plates and incubated at 30°C for 4 days. (C) Terminal phenotype of Δ_pfh1_ cells. Cells that germinated from single Δ_pfh1_ spores on a YE plate were photographed. (D) DAPI staining of germinating Δ_pfh1_ cells. Δ_pfh1_ spores derived form pfh1 + /pfh1::ura4 + were preferentially germinated in minimal medium lacking uracil. Germinating cells cultured for 18 h were fixed with 70% ethanol and stained with DAPI. (E) Flow cytometric analysis of pfh1 null cells after spore germination. Spores derived from pfh1 + /pfh1::ura4 + and ura4 + /ura4-D18 control strain were germinated in minimal medium lacking uracil at 30°C. Germinating cells were collected every 2 h, fixed with 70% ethanol and analyzed by flow cytometry. The positions of the 1C and 2C DNA peaks are indicated.
Figure 6
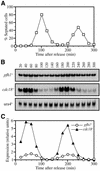
Cell cycle northern blot analysis of the pfh1 + gene. h – cdc25-22 cells were arrested in late G2 and then released to the permissive temperature. Cell aliquots were taken every 20 min. The growth of the synchronized cells was followed for two generations. (A) The cell cycle profile was monitored by measuring the percentage of septated cells at each time point. (B) The expression of pfh1 +, cdc18 + and ura4 + were examined by northern hybridization. (C) The relative mRNA levels of pfh1 + and cdc18 +. The mRNA level of pfh1 +, cdc18 + and ura4 + shown in (B) were quantified using BAS2000 (Fuji Film). The relative ratio against ura4+ was calculated and plotted. The lowest signals were adjusted as one relative unit.
Figure 7
Purification of recombinant wild-type and mutant Pfh1 proteins. Crude extracts prepared from uninduced (lanes 1, 6, 11 and 16) and induced (lanes 2, 7, 12 and 17) E.coli BL21 cells harboring pET43-Pfh1 (lanes 1–5 and 11–15) and pET43-Pfh1K338E (lanes 6–10 and 16–20) were subjected to 8% SDS–PAGE along with fractions from each purification step as indicated, and the gel was either Coomassie stained (lanes 1–10) or analyzed in western blot analyses (lanes 11–20) using monoclonal antibody specific for penta-histidine (α-His; Qiagen). The numbers on the left indicate the molecular sizes (in kDa) of marker proteins (M; Bio-Rad). M, molecular weight marker; CE(–), crude extracts (20 µg) prepared from uninduced E.coli cells; CE(+), crude extracts (20 µg) prepared from induced E.coli cells; Ni2+, fractions (1 µg for lanes 3 and 8; 400 ng for lanes 13 and 18) eluted from the Ni2+-charged HiTrap-chelating column; Hep, fractions (500 ng for lanes 4 and 9; 200 ng for lanes 14 and 19) from the HiTrap heparin column; S 200, fractions (500 ng for lanes 5 and 10; 200 ng for lanes 15 and 20) obtained from the Superdex 200 column. Arrowheads indicate the position of recombinant wild-type or mutant NusA-Pfh1 proteins.
Figure 8
Hydrolysis of ATP and unwinding of duplex DNA by Pfh1 enzymes. (A) DNA-dependent ATPase activities of wild-type Pfh1 and mutant Pfh1K338E were measured as described in Materials and Methods. The amounts of enzymes (Wt, NusA-Pfh1; mutant, NusA-Pfh1K338E) added and omissions of M13 sscDNA (–ssDNA) and Mg2+ (–Mg2+) were as indicated. The amounts of ATP hydrolyzed are presented at the bottom. (B) Helicase activities of NusA-Pfh1 and NusA-Pfh1K338E were measured as described in Materials and Methods. The reactions were incubated at 37°C for 10 min. The controls without ATP (–ATP) or MgCl2 (–Mg2+) were as indicated. The structure of the partial duplex ΦX174 sscDNA substrate used was shown. The asterisks indicate 32P-labeled ends. An arrow indicates the position where the labeled oligonucleotides migrated. The amounts of substrate unwound are presented at the bottom. (C) Schematic structures of substrates used are shown at the top. The asterisks indicate 32P-labeled ends. The indicated amounts of NusA-Pfh1 in a 20 µl reaction mixture were incubated with 15 fmol of either 5′- or 3′-overhang 21-bp duplex DNA substrate at 37°C for 10 min. The products were analyzed on a 12% polyacrylamide gel. Lanes labeled B denote boiled substrate controls. An arrow indicates the position where the labeled oligonucleotides migrated. The amounts of substrate unwound are presented at the bottom.
Figure 9
Nuclear localization of Pfh1-GFP. Exponentially growing S.pombe cells expressing Pfh1-GFP under the control of the pfh1 + promoter (left) and wild-type cells (right) in minimal medium were analyzed by fluorescent microscopy.
Similar articles
- Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast.
Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS. Ryu GH, et al. Nucleic Acids Res. 2004 Aug 9;32(14):4205-16. doi: 10.1093/nar/gkh720. Print 2004. Nucleic Acids Res. 2004. PMID: 15302919 Free PMC article. - Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2-Cdc24 complex to DNA polymerase delta.
Tanaka H, Ryu GH, Seo YS, MacNeill SA. Tanaka H, et al. Nucleic Acids Res. 2004 Dec 2;32(21):6367-77. doi: 10.1093/nar/gkh963. Print 2004. Nucleic Acids Res. 2004. PMID: 15576681 Free PMC article. - Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates.
Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Morishita T, et al. Mol Cell Biol. 2005 Sep;25(18):8074-83. doi: 10.1128/MCB.25.18.8074-8083.2005. Mol Cell Biol. 2005. PMID: 16135799 Free PMC article. - DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase.
Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. Sabouri N, et al. Genes Dev. 2012 Mar 15;26(6):581-93. doi: 10.1101/gad.184697.111. Genes Dev. 2012. PMID: 22426534 Free PMC article. - The functions of the multi-tasking Pfh1Pif1 helicase.
Sabouri N. Sabouri N. Curr Genet. 2017 Aug;63(4):621-626. doi: 10.1007/s00294-016-0675-2. Epub 2017 Jan 4. Curr Genet. 2017. PMID: 28054200 Free PMC article. Review.
Cited by
- Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast.
Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS. Ryu GH, et al. Nucleic Acids Res. 2004 Aug 9;32(14):4205-16. doi: 10.1093/nar/gkh720. Print 2004. Nucleic Acids Res. 2004. PMID: 15302919 Free PMC article. - The TPR-containing domain within Est1 homologs exhibits species-specific roles in telomerase interaction and telomere length homeostasis.
Sealey DC, Kostic AD, LeBel C, Pryde F, Harrington L. Sealey DC, et al. BMC Mol Biol. 2011 Oct 18;12:45. doi: 10.1186/1471-2199-12-45. BMC Mol Biol. 2011. PMID: 22011238 Free PMC article. - Identification of cell cycle-regulated genes in fission yeast.
Peng X, Karuturi RK, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, Liu J. Peng X, et al. Mol Biol Cell. 2005 Mar;16(3):1026-42. doi: 10.1091/mbc.e04-04-0299. Epub 2004 Dec 22. Mol Biol Cell. 2005. PMID: 15616197 Free PMC article. - Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Dehghani-Tafti S, Levdikov V, Antson AA, Bax B, Sanders CM. Dehghani-Tafti S, et al. Nucleic Acids Res. 2019 Apr 8;47(6):3208-3222. doi: 10.1093/nar/gkz028. Nucleic Acids Res. 2019. PMID: 30698796 Free PMC article. - The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA.
Pinter SF, Aubert SD, Zakian VA. Pinter SF, et al. Mol Cell Biol. 2008 Nov;28(21):6594-608. doi: 10.1128/MCB.00191-08. Epub 2008 Aug 25. Mol Cell Biol. 2008. PMID: 18725402 Free PMC article.
References
- MacNeill S.A. and Burgers,P.M.J. (2000) Chromosomal DNA replication in yeast: enzymes and mechanisms. In Fantes,P. and Beggs,J. (eds), The Yeast Nucleus. Oxford University Press, Oxford, UK, pp. 19–57.
- Nasmyth K. and Nurse,P. (1981) Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases