Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect - PubMed (original) (raw)
Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect
Catherine A Blankenship et al. J Virol. 2002 Dec.
Abstract
The human cytomegalovirus IRS1 and TRS1 open reading frames encode immediate-early proteins with identical N-terminal domains and divergent C-terminal regions. Both proteins have been shown previously to activate reporter genes in transfection assays in cooperation with other viral gene products. We have constructed two viruses carrying substitution mutations within either the IRS1 or TRS1 open reading frame. ADsubIRS1 failed to produce the related IRS1 and IRS1(263) proteins, but it replicated with normal kinetics to produce a wild-type yield in human fibroblasts. The addition in trans of the IRS1(263) protein, which antagonizes the ability of IRS1 and TRS1 proteins to activate reporter genes, did not inhibit the growth of the mutant virus. ADsubTRS1 failed to produce the TRS1 protein, and it generated an approximately 200-fold-reduced yield of infectious virus in comparison to its wild-type parent. Viral DNA accumulated normally, as did a set of viral mRNAs that were monitored in ADsubTRS1-infected cells. However, two tegument proteins were partially mislocalized and infectious virus particles did not accumulate to normal levels within ADsubTRS1-infected cells. Further, infectious ADsubTRS1 particles sedimented abnormally in a glycerol-tartrate gradient, indicating that the structure of the mutant particles is aberrant. Our analysis of the ADsubTRS1 phenotype indicates that the TRS1 protein is required, either directly or indirectly, for efficient assembly of virus particles.
Figures
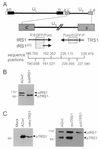
FIG. 1.
Locations of alterations in mutant and revertant viruses and effect of mutations on protein expression within infected cells. (A) Diagram of IRS1 and TRS1 coding regions depicting the locations of mutations. The top portion displays the entire viral genome with unique (UL and US) and repeated (a, b, and c) domains. The region containing the IRS1, IRS1263, and TRS1 open reading frames (arrows) is expanded and displayed below. Cross-hatched boxes, regions deleted when producing the substitution mutations in AD_sub_IRS1 and AD_sub_TRS1. Above the boxes the inserted marker cassette is indicated (P, SV40 early promoter; EGFP, EGFP coding region; Puro, puromycin resistance gene). Bar on TRS1 arrow, single base pair change (C to A) introduced into AD_rev_TRS1. The sequence boundaries of the open reading frames and deletions are indicated at the bottom. (B) Western blot analysis of pIRS1 and pTRS1 expression after infection at a multiplicity of infection of 5 PFU/cell with AD_wt_ or AD_sub_IRS1. (C) (Left) Western blot assay of pTRS1 expression after infection at a multiplicity of infection of 5 PFU/cell with AD_wt_ or at a multiplicity of infection of 3 transducing units/cell with retroTRS1. (Right) Western blot assay of pIRS1 and pTRS1 expression in retroTRS1-containing cells infected with AD_sub_TRS1 or AD_rev_TRS1 at a multiplicity of infection of 5 PFU/cell.
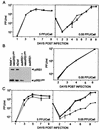
FIG. 2.
Production of infectious progeny in the medium of cultures infected with wild-type, mutant, or revertant viruses. HCMV was used to infect human fibroblasts at the indicated multiplicities of infection, and retroviruses were used at a multiplicity of infection of 3 transducing units/cell. Virus that accumulated in the medium of infected cultures was determined by plaque assays on fibroblasts (AD_wt_ and AD_sub_IRS1) and retroTRS1-infected fibroblasts (AD_sub_TRS1 and AD_rev_TRS1) in duplicate. (A) Growth of AD_wt_ (♦) and AD_sub_IRS1 (▪). (B) (Left) Western blot assay of pIRS1 and pIRS1263 expression after infection with AD_wt_ or AD_sub_IRS1 alone or in combination with retroIRS263; (right) growth of AD_wt_ (♦), AD_sub_IRS1 (▪), AD_wt_ plus retroIRS1263 (▴), and AD_sub_TRS1 plus retroIRS1263 (×). (C) Growth of AD_wt_ (♦), AD_rev_TRS1 (▪), AD_sub_TRS1 (▴), and AD_sub_TRS1 plus retroTRS1 (×).
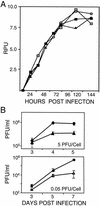
FIG. 3.
Accumulation of DNA and intracellular virus after infection with wild-type or mutant virus. (A) DNA accumulation. Human fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with AD_wt_ (), AD_sub_IRS1 (igwidth>), AD_sub_TRS1 (▪), or AD_sub_TRS1 plus retroTRS1 (igwidth>). Total-infected-cell DNA was prepared at the indicated times, and viral DNA was assayed by Southern blotting with a 32P-labeled HCMV genomic probe DNA. Radioactivity was quantified with a phosphorimager, and results are presented as relative phosphorimager units (RPU). (B) Intracellular virus accumulation. Cells were infected at the indicated multiplicities of infection with AD_wt_ (♦) or AD_sub_TRS1 (▴ or igwidth>). Cells were harvested at the indicated times and freeze-thawed twice, and infectious virus was quantified by plaque assay on retroTRS1-infected fibroblasts. Samples were assayed in duplicate.
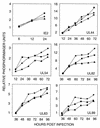
FIG. 4.
Accumulation of viral mRNAs after infection with wild-type or mutant virus. Fibroblasts were infected at a multiplicity of infection of 0.1 PFU/cell with AD_wt_ (♦), AD_sub_IRS1 (▪), or AD_sub_TRS1 (▴). Total cellular RNA was prepared at the indicated times, viral mRNAs were assayed by Western blotting with 32P-labeled probe DNAs, and radioactivity relative to that for a cellular mRNA (cPLA2) was quantified with a phosphorimager. Similar results were obtained in two independent experiments.
FIG. 5.
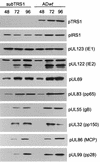
Accumulation of viral proteins after infection with wild-type or mutant virus. Fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with AD_wt_ or AD_sub_TRS1. Cell extracts were prepared at the indicated times (hours postinfection), and viral proteins were assayed by Western blotting with antibodies specific for the indicated proteins. gB, glycoprotein B; MCP, major capsid protein.
FIG. 6.
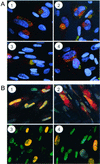
Localization of late viral proteins in infected cells. Fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with AD_wt_ (1 and 2) or AD_sub_TRS1 (3 and 4). Cells were fixed 60 (1 and 3) or 72 h (2 and 4) later and treated with RNase A. pUL99 (A) was visualized by using a specific monoclonal antibody plus a fluorescein-conjugated secondary antibody (green); Golgi bodies were stained with an antibody to a Golgi-specific marker (mannosidase II) plus a Cy5-conjugated secondary antibody (red), and DNA was stained with Hoechst 33342 dye (blue). Limited colocalization of pUL99 and mannosidase II produces a yellow signal. pUL83 (B) was detected with a specific antibody plus a Cy5-conjugated secondary antibody (red), and DNA was stained with YOYO-1 (green). Colocalization of pUL83 and nuclear DNA yields a yellow signal.
FIG. 7.
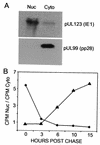
Kinetic analysis of pUL83 location after synthesis in infected cells. (A) Fractionation of cells. Fibroblasts were infected with AD_wt_ at a multiplicity of infection of 0.05 PFU/cell. At 72 h later, cells were subjected to mechanical fractionation in hypotonic buffer, separating nuclear (Nuc) and cytoplasmic (Cyto) fractions. The locations of the nuclear pUL123 (IE1) and cytoplasmic pUL99 viral proteins were ascertained by Western blot assay. (B) Localization of pUL83. Fibroblasts were infected at a multiplicity of infection of 0.05 PFU/cell with AD_wt_ (♦) or AD_sub_TRS1 (▴). At 65 h later, cells were labeled with [35S]methionine plus [35S]cysteine for 10 min, washed, and then chased in the presence of unlabeled methionine plus cysteine. Lysates were prepared at the indicated times, and immunoprecipitated pUL83 from nuclear and cytoplasmic fractions was analyzed by electrophoresis. The radioactivity in pUL83-specific bands was quantified with a phosphorimager, and the ratio of 35S-labeled pUL83 in the nucleus to that in the cytoplasm was calculated.
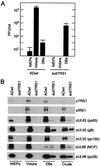
FIG. 8.
Infectivity and protein constituents of wild-type and mutant virus particles. Virus particles from the medium of fibroblasts infected with AD_wt_ or AD_sub_TRS1 were partially purified by sedimentation through a sorbitol cushion to produce partially purified particles (crude). The particles in the crude fraction were further purified by sedimentation in a glycerol-tartrate gradient. Virions, NIEPs, and (DBs) were separated on the gradient containing the AD_wt_ preparation. No virions or NIEPs were evident in the AD_sub_TRS1 preparation, and the regions of the gradient that would normally contain these particles were collected in blind fashion and processed. (A) The infectivity present in fractions from glycerol-tartrate gradients was analyzed by plaque assay on retroTRS1-infected fibroblasts. Three independent experiments were performed, and the averages plus standard deviations are displayed. (B) Viral proteins in gradient fractions were analyzed by Western blotting with antibodies to the indicated proteins. gB and MCP are as defined for Fig. 5.
Similar articles
- Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids.
Adamo JE, Schröer J, Shenk T. Adamo JE, et al. J Virol. 2004 Oct;78(19):10221-9. doi: 10.1128/JVI.78.19.10221-10229.2004. J Virol. 2004. PMID: 15367587 Free PMC article. - Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2.
Stasiak PC, Mocarski ES. Stasiak PC, et al. J Virol. 1992 Feb;66(2):1050-8. doi: 10.1128/JVI.66.2.1050-1058.1992. J Virol. 1992. PMID: 1370547 Free PMC article. - Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis.
Pari GS, Kacica MA, Anders DG. Pari GS, et al. J Virol. 1993 May;67(5):2575-82. doi: 10.1128/JVI.67.5.2575-2582.1993. J Virol. 1993. PMID: 8386266 Free PMC article. - Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci.
Colberg-Poley AM. Colberg-Poley AM. Intervirology. 1996;39(5-6):350-60. doi: 10.1159/000150506. Intervirology. 1996. PMID: 9130045 Review. - Human cytomegalovirus phosphoproteins and glycoproteins and their coding regions.
Jahn G, Mach M. Jahn G, et al. Curr Top Microbiol Immunol. 1990;154:171-85. doi: 10.1007/978-3-642-74980-3_7. Curr Top Microbiol Immunol. 1990. PMID: 2161320 Review. No abstract available.
Cited by
- Antagonism of Protein Kinase R by Large DNA Viruses.
Olson AT, Child SJ, Geballe AP. Olson AT, et al. Pathogens. 2022 Jul 12;11(7):790. doi: 10.3390/pathogens11070790. Pathogens. 2022. PMID: 35890034 Free PMC article. Review. - Human Cytomegalovirus pTRS1 and pIRS1 Antagonize Protein Kinase R To Facilitate Virus Replication.
Ziehr B, Vincent HA, Moorman NJ. Ziehr B, et al. J Virol. 2016 Mar 28;90(8):3839-3848. doi: 10.1128/JVI.02714-15. Print 2016 Apr. J Virol. 2016. PMID: 26819306 Free PMC article. - Essential role of protein kinase R antagonism by TRS1 in human cytomegalovirus replication.
Braggin JE, Child SJ, Geballe AP. Braggin JE, et al. Virology. 2016 Feb;489:75-85. doi: 10.1016/j.virol.2015.11.032. Epub 2015 Dec 21. Virology. 2016. PMID: 26716879 Free PMC article. - Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins.
Mouna L, Hernandez E, Bonte D, Brost R, Amazit L, Delgui LR, Brune W, Geballe AP, Beau I, Esclatine A. Mouna L, et al. Autophagy. 2016;12(2):327-42. doi: 10.1080/15548627.2015.1125071. Autophagy. 2016. PMID: 26654401 Free PMC article. - Functional annotation of human cytomegalovirus gene products: an update.
Van Damme E, Van Loock M. Van Damme E, et al. Front Microbiol. 2014 May 19;5:218. doi: 10.3389/fmicb.2014.00218. eCollection 2014. Front Microbiol. 2014. PMID: 24904534 Free PMC article. Review.
References
- Chee, M. S., et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. - PubMed
- Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources