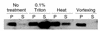A novel dispersin protein in enteroaggregative Escherichia coli - PubMed (original) (raw)
A novel dispersin protein in enteroaggregative Escherichia coli
Jalaluddin Sheikh et al. J Clin Invest. 2002 Nov.
Abstract
Enteroaggregative Escherichia coli (EAEC) is a diarrheal pathogen defined by its characteristic aggregative adherence (AA) to HEp-2 cells in culture. We have previously shown that EAEC strains secrete a 10-kDa protein that is immunogenic in a human EAEC challenge model. We report here that this protein is encoded by a gene (called aap) lying immediately upstream of that encoding the AggR transcriptional activator, and that aap is under AggR control. The product of aap has a typical signal sequence and is secreted to the extracellular milieu, where it remains noncovalently attached to the surface of the bacterium. EAEC aap mutants aggregate more intensely than the wild-type parent in a number of assays, forming larger aggregates and fewer individual bacteria. Infection of colonic biopsies with wild-type EAEC strain 042 and its aap mutant revealed more dramatic autoagglutination of the mutant compared with the wild-type parent. Our data suggest that the aap gene product participates in formation of a surface coat that acts to disperse the bacteria, thus partially counteracting aggregation mediated by aggregative adherence fimbriae. We have therefore named the aap gene product "dispersin," and we propose that it may be representative of a functional class of colonization factors. Since dispersin is expressed in vivo, is highly immunogenic, and is present in most EAEC strains, it holds considerable promise as an EAEC immunogen.
Figures
Figure 1
Map of aap-aggR loci from strains 042 (producing AAF/II) and 17-2 (producing AAF/I). The aap-aggR locus is highly conserved between the two strains, despite divergence of the fimbrial loci. The aap-aggR intergenic region is 99% identical in the two strains, except for a 50-nucleotide insertion present in the 17-2 sequence. IS, insertion sequence. Values percent indicate nucleotide homology between 042 and 17-2 sequences.
Figure 2
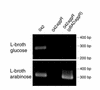
RT-PCR for aap transcript in 042, 042_aggR_, and 042_aggR_(pBAD_aggR_). Cultures were grown to late log phase in either L-broth with 0.5% glucose or L-broth with 0.5% arabinose. RT-PCR was performed as described in Methods, and products were separated on 1% agarose gels. The aap transcript migrates as a 0.3-kb species as predicted.
Figure 3
Localization of the aap gene product. Western immunoblot analysis was performed on 042 grown in L-broth and subjected to addition of 0.1% Triton X-100 detergent, incubation at 60°C for 20 minutes, or vigorous vortexing for 10 minutes. Cultures were grown to late log phase, and bacteria were pelleted by centrifugation. Pellets (P) were prepared by boiling whole pellet in SDS-PAGE buffer and separating 10% of the sample derived from a 1.0-ml culture. Supernatant (S) was prepared by TCA precipitation. Western immunoblots were performed by standard methods using highly specific polyclonal antiserum to Aap protein. Aap migrates at the predicted 10-kDa size.
Figure 4
Electron microscopy of 042 and mutants. (a) Immunogold transmission electron microscopy for Aap protein. Black arrows indicate surface protein detected by anti-Aap antibody. Strains examined by scanning electron microscopy are 042 (b and c), 042_aafA_ (d), 042_aap_ (e–g), and 042_aap_(pAap) (h). In i, 042_aap_ was incubated with 3 mg/ml purified Aap protein prior to fixing and staining. White arrows indicate AAF fimbriae in all specimens. All bars indicate 1 μm.
Figure 5
Autoagglutination of 042 and mutants in L-broth. L-broth cultures were grown overnight at 37°C with shaking, then permitted to settle undisturbed at room temperature for 1 hour. (a) Broth cultures revealing a pellet at the bottom of the culture tube correlating with autoagglutination of bacterial cells. (b) Transmission electron microscopy of 042 and 042_aap_ cultures. Ten microliters of L-broth cultures from a were withdrawn prior to incubation on the bench-top and processed for negative staining. Bars, 1 μm.
Figure 6
Spontaneous settling of 042 and mutants. Overnight L-broth cultures containing 0.2 mM IPTG were permitted to settle undisturbed at room temperature. At 15-minute intervals, a 100-μl sample was withdrawn from the top of each broth culture and the OD600 of the withdrawn sample was determined. Rate of clearing of the uppermost portion of the culture correlates with rate of spontaneous settling, a measure of agglutination. Samples were tested in triplicate. Bars represent SEM.
Figure 7
Role of aap in epithelial cell adherence. Assays were performed for 3 hours in DMEM–0.5% glucose with 0.2 mM IPTG at 37°C in atmosphere. (a) 042; (b) 042_aap_; (c) 042_aap_(pAap). (d and e) The standard HEp-2 cell adherence assay was performed using strain 042_aap_, but with pure Aap (d) or pure BSA (e) added concurrently with the bacterial inoculum. Each protein was added to a final concentration of 0.1 mg/ml. (f) Quantitation of HEp-2 cell adherence on confluent monolayers growing in DMEM without IPTG (042 and 042_aap_) or with indicated IPTG concentration [042_aap_(pAap)]. Bacteria were removed from glass using 1% Triton X-100 and quantitated by viable count. Assays were performed in triplicate, and means are illustrated in histograms with error bars indicating SE. *P < 0.05 vs. 042 by one-tailed t test of log-transformed data; **P < 0.05 vs. 042_aap_ by one-tailed t test of log-transformed data.
Figure 8
Glass adherence assay. (a–c) Assay was performed for 3 hours in DMEM–0.5% glucose with 0.1 mM IPTG at 37°C in atmosphere. (a) 042; (b) 042_aap_; (c) 042_aap_(pAap). (d) Quantitation of adherence grown in DMEM without IPTG (042 and 042_aap_) or with the indicated concentration of IPTG [(042_aap_(pAap)]. Bacteria were removed from glass using 1% Triton X-100 and quantitated by viable count. Assays were performed in triplicate, and means are illustrated in histograms with error bars indicating SEM. *P < 0.05 vs. 042_aap_ by one-tailed t test of log-transformed data.
Figure 9
In vitro organ culture assay of 042 (a) and 042_aap_ (b). Bacterial cultures were incubated with colonic biopsies from normal children for 6 hours; then specimens were fixed, stained, and examined by scanning electron microscopy. White arrows (a) denote loose adherence of 042; gray arrows (b) show tight aggregates typical of 042_aap_. Bars, 10 μm.
Figure 10
Penetration of 042 and 042_aap_ through mucus. Columns of 10 mg/ml mucin in HBSS were prepared in tuberculin syringes, and the bacterial inocula were applied to the top of the column. The columns were incubated at 37°C for 75 minutes. After this time, the mucin suspension was drained from the column in 0.1-ml fractions, and the number of 042 (black bars) or 042_aap_ (white bars) was determined by quantitative plating in triplicate. Bars represent SD. Fraction 10 represents the bottom of the column, fraction 1 the top. *P < 0.05 by Student’s t test of log-transformed data.
Figure 11
Western immunoblots for Aap performed on supernatants (S) or pellets (P) of overnight L-broth cultures with or without 0.1% Triton X-100. Positions of molecular weight markers are shown at right.
Similar articles
- Aggregative Adherence and Intestinal Colonization by Enteroaggregative Escherichia coli Are Produced by Interactions among Multiple Surface Factors.
Blanton LV, Wang LT, Hofmann J, DuBow J, Lafrance A, Kwak S, Bowers L, Levine MA, Hale CO, Meneely PM, Okeke IN. Blanton LV, et al. mSphere. 2018 Mar 21;3(2):e00078-18. doi: 10.1128/mSphere.00078-18. eCollection 2018 Mar-Apr. mSphere. 2018. PMID: 29577084 Free PMC article. - The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative E. coli 042.
Imuta N, Nishi J, Tokuda K, Fujiyama R, Manago K, Iwashita M, Sarantuya J, Kawano Y. Imuta N, et al. Infect Immun. 2008 Mar;76(3):1247-56. doi: 10.1128/IAI.00758-07. Epub 2007 Dec 26. Infect Immun. 2008. PMID: 18160483 Free PMC article. - A Mini-Review of Enteroaggregative Escherichia coli with a Specific Target on the Virulence Factors Controlled by the AggR Master Regulator.
Izquierdo-Vega JA, Castillo-Juarez RJ, Sánchez-Gutiérrez M, Ares MA, De La Cruz MA. Izquierdo-Vega JA, et al. Pol J Microbiol. 2023 Dec 16;72(4):347-354. doi: 10.33073/pjm-2023-037. eCollection 2023 Dec 1. Pol J Microbiol. 2023. PMID: 37875068 Free PMC article. Review. - The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli.
Monteiro BT, Campos LC, Sircili MP, Franzolin MR, Bevilacqua LF, Nataro JP, Elias WP. Monteiro BT, et al. Diagn Microbiol Infect Dis. 2009 Sep;65(1):81-4. doi: 10.1016/j.diagmicrobio.2009.05.011. Diagn Microbiol Infect Dis. 2009. PMID: 19679242 - Enteroaggregative Escherichia coli pathogenesis.
Nataro JP. Nataro JP. Curr Opin Gastroenterol. 2005 Jan;21(1):4-8. Curr Opin Gastroenterol. 2005. PMID: 15687877 Review.
Cited by
- Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli.
Jønsson R, Struve C, Boisen N, Mateiu RV, Santiago AE, Jenssen H, Nataro JP, Krogfelt KA. Jønsson R, et al. Infect Immun. 2015 Apr;83(4):1396-405. doi: 10.1128/IAI.02820-14. Epub 2015 Jan 26. Infect Immun. 2015. PMID: 25624357 Free PMC article. - Rise of the microbes.
Mahan MJ, Kubicek-Sutherland JZ, Heithoff DM. Mahan MJ, et al. Virulence. 2013 Apr 1;4(3):213-22. doi: 10.4161/viru.23380. Epub 2013 Jan 18. Virulence. 2013. PMID: 23334178 Free PMC article. Review. - Antibiofilm agents with therapeutic potential against enteroaggregative Escherichia coli.
Kwasi DA, Babalola CP, Olubiyi OO, Hoffmann J, Uzochukwu IC, Okeke IN. Kwasi DA, et al. PLoS Negl Trop Dis. 2022 Oct 6;16(10):e0010809. doi: 10.1371/journal.pntd.0010809. eCollection 2022 Oct. PLoS Negl Trop Dis. 2022. PMID: 36201560 Free PMC article. - Evaluation of a multiplex PCR for identification of enteroaggregative Escherichia coli.
Cordeiro F, da Silva Gomes Pereira D, Rocha M, Asensi MD, Elias WP, Campos LC. Cordeiro F, et al. J Clin Microbiol. 2008 Feb;46(2):828-9. doi: 10.1128/JCM.01865-07. Epub 2007 Dec 12. J Clin Microbiol. 2008. PMID: 18077638 Free PMC article. No abstract available. - New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family.
Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. Boisen N, et al. Infect Immun. 2008 Jul;76(7):3281-92. doi: 10.1128/IAI.01646-07. Epub 2008 Apr 28. Infect Immun. 2008. PMID: 18443096 Free PMC article.
References
- Okeke IN, Nataro JP. Enteroaggregative Escherichia coli. Lancet Infect Dis. 2001;1:304–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous