Detection of peptides, proteins, and drugs that selectively interact with protein targets - PubMed (original) (raw)
Detection of peptides, proteins, and drugs that selectively interact with protein targets
Ilya G Serebriiskii et al. Genome Res. 2002 Nov.
Abstract
Genome sequencing has been completed for multiple organisms, and pilot proteomic analyses reported for yeast and higher eukaryotes. This work has emphasized the facts that proteins are frequently engaged in multiple interactions, and that governance of protein interaction specificity is a primary means of regulating biological systems. In particular, the ability to deconvolute complex protein interaction networks to identify which interactions govern specific signaling pathways requires the generation of biological tools that allow the distinction of critical from noncritical interactions. We report the application of an enhanced Dual Bait two-hybrid system to allow detection and manipulation of highly specific protein-protein interactions. We summarize the use of this system to detect proteins and peptides that target well-defined specific motifs in larger protein structures, to facilitate rapid identification of specific interactors from a pool of putative interacting proteins obtained in a library screen, and to score specific drug-mediated disruption of protein-protein interaction.
Figures
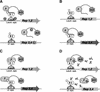
Figure 1
Outline of Dual Bait System. (A) An activation domain-fused prey (P) interacts with a LexA-fused bait (B1) to drive transcription of _lexAop_-responsive LEU2 and lacZ reporters, but does not interact with a cI-fused bait (B2) and, thus, does not turn on transcription of _cIop_-responsive LYS2 and gusA reporters. The prey may represent a protein, as in Applications 1 and 3, or a peptide aptamer, as in Application 2. Note, in this example, the cI-Bait is drawn as representing a negative control for prey binding; the system can also be configured so that prey interacts with either or both baits, as in Serebriiskii et al. (1999). Points addressed in optimization of the Dual Bait two-hybrid system in this study (Table 1) are as follows: (1), varying expression level of baits; (2), enriching polylinkers to facilitate cloning of baits; (3), varying sensitivity of reporters; (4), diversifying plasmid antibiotic markers to facilitate isolation of library plasmid in E. coli. In addition, we have developed a robust yeast strain, SKY473, which is suitable both for bait testing and interaction mating in this system. (B) As described in Application 3, preys isolated against LexA-B1 and counterselected against cI-B2 are subsequently challenged with cI-B1 and LexA-B2, in a bait swap experiment. Those preys binding specifically to the B1 domain (as opposed to B1 specifically in the context of a LexA-B1 fusion protein) are retained preferentially. (C) One protein (P) may use different surface motifs to bind two different partners (B1, B2). (D) Application of a small molecule (drug or peptide) that obstructs one of the interactions shown in C will selectively turn off two of the four reporter genes, allowing subtractive scoring.
Figure 2
Targeting of interacting proteins or peptides to small sequence motifs. (A) Differential interaction of Cdc42 effectors as detected by the Dual Bait procedure. Two Cdc42-interacting proteins, FBP17 (Fuchs et al. 2001) and SPEC1 (Pirone et al. 2000) were isolated by a yeast two-hybrid screen with LexA-Cdc42-L28. In this screen, 107 diploids arising from an interaction mating (Finley and Brent 1994)-based screen yielded 25 positive colonies showing interactions with LexA-Cdc42 L28. Counterscreening against cI fused to activated Cdc42 lacking the insert region (Cdc42 L28-Δ8) revealed two clones, both containing the same cDNA, showing differential binding. All yeast shown contain both baits, and AD-fusions as indicated at left. Beside AD-fused FBP17 (specific for Cdc42-L28) and SPEC1 (bound both forms equivalently), AD-Pak1, which binds both forms of Cdc42, and AD-Pak1 LL, which cannot bind either form of Cdc42 (Sells et al. 1997), are included as controls. Results with growth and colorimetric reporters for Cdc42-L28-Δ8 [gusA (X-Gluc) and LYS2, first and third lines] and LexA-Cdc42-L28 [lacZ (X-Gal) and LEU2, second and fourth lines], are shown. (B) Selection of peptides specifically targeting HEF1-DLVD. A LexA-HEF1(DLVD) bait was used to identify interacting peptide aptamers, in collaboration with R. Finley. Selected peptides were rescreened in parallel against LexA-HEF1(DVLD) or cI-HEF1(DLVA), differing from the original bait by only a D363A substitution. Shown are peptides that interact with both DLVD and DLVA variants (DLVD = DLVA; activates all four reporters; representative sequence HHASTPRRESPGIMSPL); or with DLVD but not DLVA (DLVD>>DLVA; activates lacZ and LEU2 only; representative sequence, SGKFGEALPGWLSSACWCFG), and a nonspecific control peptide (negative; activates no reporters; representative sequence, EQLKYNRFWPWQWWGGRRLR). (C) Confirmation that peptides interact with HEF1 in an in vitro system, using pulldowns of endogenous HEF1 from cell lysates with the three GST-fused peptides from B, or with GST only. (Top) Levels of GST peptide or GST only in reaction; (bottom) associated HEF1.
Figure 3
Use of Dual Bait reagents to reduce false positive background. (Top) The RBΔ663 and RBΔex22 mutants of pRB are described in Sellers et al. (1998), and were expressed in the context of a large pocket domain of pRB containing amino acids from 379 to 928. SKY48 yeast expressing LexA-RBΔ663 and cI-RBΔex22 baits were used to screen three different libraries. (Lines 1,2) Numbers of clones positive for LexA-RBΔ663-responsive (LEU2, lacZ) but negative for cI-RBΔex22-responsive (gusA) reporters. (Line 3) Number of discrete genes represented among the clones. (Line 4) Number of clones positive for cI-RBΔ663-responsive (GusA), but not LexA-RBΔex22-responsive (LEU, lacZ) reporters; note, this reduction from line 2 values was not observed with retransformation testing with the original LexA-RBΔ663 and cI-RBΔex22 baits. (Line 5) Number of genes represented in line 4 set of clones. (Line 6) Number of surviving genes that were validated by coimmunoprecipiation and other techniques. (*) Note, papillomavirus E7 was identified from two different fetal brain libraries with B42 or GAL4 as activation domains. Although a legitimate pRB interactor, as E7 is not normally expressed in brain, it may represent an artifact of the libraries' construction. (Bottom) HA-tagged E7 and pRB derivatives were overexpressed in Saos-2 (_Rb_−/−) osteosarcoma cells and their interaction was determined by immunoprecipitation with anti-HA antibody (HA 11, BAbCO). The precipitated proteins pRB, pRBΔ663, and RBΔex22 (RBΔ22 in figure) were detected by immunoblotting with anti-RB antibody (XZ56). Input proteins are 10% of that used in immunoprecipitation.
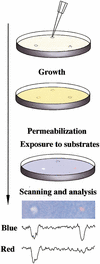
Figure 4
Drug disruption of protein–protein interactions. (Top) Yeast containing baits and preys are mixed with low-melt agarose and poured over appropriate dropout growth medium. After agarose is set, 1 μL of each compound to be tested or solvent negative control is dropped on the plate. Yeast are incubated for 1–2 d, then permeabilized and overlaid with Z-buffer, Magenta-Gal (a red colorimetric substrate for LacZ) and X-Gluc (a blue colorimetric substrate for GusA). (Bottom) The result shown derives from a mixed population of yeast strains containing LexA-Ras and AD-Raf or cI-Ras and AD-RalGDS. In this example, only the colorimetric (lacZ and gusA) reporters are being assessed. Fungicide (left) inhibits both lacZ and gusA signal, whereas a specific Ras–Raf interaction inhibitor reduces only gusA (blue) output, leaving a red spot; solvent control produced no spots (data not shown). Shown below the spots are results obtained following a scan of plate, import of image into NIH Image, and performance of densitometry for signal intensity in blue versus red across the spot midline.
Similar articles
- Analysis of protein-protein interactions utilizing dual bait yeast two-hybrid system.
Serebriiskii IG, Kotova E. Serebriiskii IG, et al. Methods Mol Biol. 2004;261:263-96. doi: 10.1385/1-59259-762-9:263. Methods Mol Biol. 2004. PMID: 15064464 Review. - Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20.
Winters MJ, Pryciak PM. Winters MJ, et al. Mol Cell Biol. 2005 Mar;25(6):2177-90. doi: 10.1128/MCB.25.6.2177-2190.2005. Mol Cell Biol. 2005. PMID: 15743816 Free PMC article. - Pxl1p, a paxillin-like protein in Saccharomyces cerevisiae, may coordinate Cdc42p and Rho1p functions during polarized growth.
Gao XD, Caviston JP, Tcheperegine SE, Bi E. Gao XD, et al. Mol Biol Cell. 2004 Sep;15(9):3977-85. doi: 10.1091/mbc.e04-01-0079. Epub 2004 Jun 23. Mol Biol Cell. 2004. PMID: 15215315 Free PMC article. - Interaction trap/two-hybrid system to identify interacting proteins.
Golemis EA, Serebriiskii I, Finley RL Jr (hunt by interaction mating), Kolonin MG (hunt by interaction mating), Gyuris J, Brent R. Golemis EA, et al. Curr Protoc Cell Biol. 2011 Dec;Chapter 17:17.3.1-17.3.35. doi: 10.1002/0471143030.cb1703s53. Curr Protoc Cell Biol. 2011. PMID: 22161546 - Protein-fragment complementation assays for large-scale analysis of protein-protein interactions.
Blaszczak E, Lazarewicz N, Sudevan A, Wysocki R, Rabut G. Blaszczak E, et al. Biochem Soc Trans. 2021 Jun 30;49(3):1337-1348. doi: 10.1042/BST20201058. Biochem Soc Trans. 2021. PMID: 34156434 Free PMC article. Review.
Cited by
- Reverse yeast two-hybrid system to identify mammalian nuclear receptor residues that interact with ligands and/or antagonists.
Li H, Dou W, Padikkala E, Mani S. Li H, et al. J Vis Exp. 2013 Nov 15;(81):e51085. doi: 10.3791/51085. J Vis Exp. 2013. PMID: 24300333 Free PMC article. - Novel yeast-based strategy unveils antagonist binding regions on the nuclear xenobiotic receptor PXR.
Li H, Redinbo MR, Venkatesh M, Ekins S, Chaudhry A, Bloch N, Negassa A, Mukherjee P, Kalpana G, Mani S. Li H, et al. J Biol Chem. 2013 May 10;288(19):13655-68. doi: 10.1074/jbc.M113.455485. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525103 Free PMC article. - Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system.
Stynen B, Tournu H, Tavernier J, Van Dijck P. Stynen B, et al. Microbiol Mol Biol Rev. 2012 Jun;76(2):331-82. doi: 10.1128/MMBR.05021-11. Microbiol Mol Biol Rev. 2012. PMID: 22688816 Free PMC article. Review. - Activated alleles of the Schizosaccharomyces pombe gpa2+ Galpha gene identify residues involved in GDP-GTP exchange.
Ivey FD, Taglia FX, Yang F, Lander MM, Kelly DA, Hoffman CS. Ivey FD, et al. Eukaryot Cell. 2010 Apr;9(4):626-33. doi: 10.1128/EC.00010-10. Epub 2010 Feb 5. Eukaryot Cell. 2010. PMID: 20139237 Free PMC article. - Yeast two-hybrid, a powerful tool for systems biology.
Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U. Brückner A, et al. Int J Mol Sci. 2009 Jun 18;10(6):2763-2788. doi: 10.3390/ijms10062763. Int J Mol Sci. 2009. PMID: 19582228 Free PMC article. Review.
References
- Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. - PubMed
- Fields S, Song O. A novel genetic system to detect protein–protein interaction. Nature. 1989;340:245–246. - PubMed
- Fuchs U, Rehkamp G, Haas OA, Slany R, Konig M, Bojesen S, Bohle RM, Damm-Welk C, Ludwig J, Harbott WD, et al. The human formin-binding protein 17 (FBP17) interacts with sorting nexin, SNX2, and is an MLL-fusion partner in acute myelogeneous leukemia. Proc Natl Acad Sci. 2001;98:8756–8761. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM54168/GM/NIGMS NIH HHS/United States
- CA06927/CA/NCI NIH HHS/United States
- CA63366/CA/NCI NIH HHS/United States
- R01 CA063366/CA/NCI NIH HHS/United States
- P30 CA006927/CA/NCI NIH HHS/United States
- R01 GM054168/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases