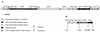The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC - PubMed (original) (raw)
The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC
I Fedtke et al. J Bacteriol. 2002 Dec.
Abstract
In Staphylococcus carnosus, the nreABC (for nitrogen regulation) genes were identified and shown to link the nitrate reductase operon (narGHJI) and the putative nitrate transporter gene narT. An nreABC deletion mutant, m1, was dramatically affected in nitrate and nitrite reduction and growth. Transcription of narT, narGHJI, and the nitrite reductase (nir) operon was severely reduced even when cells were cultivated anaerobically without nitrate or nitrite. nreABC transcripts were detected when cells were grown aerobically or anaerobically with or without nitrate or nitrite. NreA is a GAF domain-containing protein of unknown function. In vivo and in vitro studies showed that NreC is phosphorylated by NreB and that phospho-NreC specifically binds to a GC-rich palindromic sequence to enhance transcription initiation. This binding motif was found at the narGHJI, nir, and narT promoters but not at the moeB promoter. NreB is a cytosolic protein with four N-terminal cysteine residues. The second cysteine residue was shown to be important for NreB function. In vitro autophosphorylation of NreB was not affected by nitrate, nitrite, or molybdate. The nir promoter activity was iron dependent. The data provide evidence for a global regulatory system important for aerobic and anaerobic metabolism, with NreB and NreC forming a classical two-component system and NreB acting as a sensor protein with oxygen as the effector molecule.
Figures
FIG. 1.
(A) Genetic map of the genes involved in nitrite and nitrate reduction in S. carnosus. The promoters of the nir (21) and nar (24) operons and of the narT gene were mapped by primer extension analysis; the promoter upstream of nirC is postulated. Putative transcription terminators are indicated. (B) Fragments that were ligated into shuttle vector pBT2 to obtain plasmid pBT2-HOM1, used for homologous recombination to construct mutant m1. Restriction sites indicated occur naturally in the depicted genes. ′, truncation at the 3′ or 5′ end.
FIG. 2.
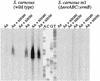
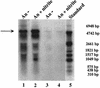
Northern blot analyses of the nreABC operon. Total RNA (15 μg) isolated from wild-type cells grown aerobically (Ae) or anaerobically (An) with (+) or without (−) nitrate (20 mM) or nitrite (2 mM), separated by gel electrophoresis, was transferred to a positively charged nylon membrane and hybridized with a digoxigenin-labeled antisense RNA fragment comprising the nreC gene. For chemiluminescent detection, CSPD (Roche) was used. The full-length transcript of the nreABC operon (approximately 2.2 kb) and the _narGHJI_-nreABC transcription unit (approximately 8.8 kb) are indicated by arrows. As a standard, 90 ng of digoxigenin-labeled RNA molecular weight marker I (Roche Molecular Biochemicals) was used. The lengths of the standard RNAs are shown in the left margin.
FIG. 3.
Nitrite reduction by S. carnosus (closed symbols) and mutant m1 (open symbols) during anaerobic growth in mBM with 500 μM nitrite. During growth, the optical density at 578 nm (OD578 nm) (triangles) and nitrite reduction (monitored as the decrease in nitrite in the growth medium) (squares) were measured.
FIG. 4.
Nitrate reduction (monitored as the decrease in nitrate in the growth medium) (squares) and nitrite accumulation in the growth medium (triangles) by wild-type S. carnosus (closed symbols) and mutant m1 (open symbols) during anaerobic growth in mBM with 1 mM nitrate.
FIG. 5.
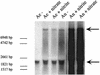
Phosphorylation of NreB and NreC. (A and B) MalE-NreB (2.9 μM, 82.5 kDa) (A) and NreB-His (2.4 μM, 41 kDa) (B) were incubated with 0.22 μM [γ-32P]ATP for 15 min (lanes 2), 30 min (lanes 3), and 45 min (lanes 4) in reaction buffer at 24°C. As a control, protein was omitted (lanes 1). (C and D) Prior to the addition of MalE-NreC (11.6 μM, 67 kDa), MalE-NreB (2.9 μM) was incubated with 20 μCi of [γ-32P]ATP in reaction buffer at 24°C for 14.5 min (first lanes). Samples (4 μl) were taken 0.5, 1, 2, 4, 10, and 15 min after addition of MalE-NreC (C) or MalE-LacZα (8.3 μM, 51 kDa) (D). (E) NreB-His (2.4 μM) was incubated in the presence of 10 μCi of [γ-32P]ATP without additions (lane 1), or with 20 mM nitrate (lane 2), 2 mM nitrite (lane 3), 0.5 mM molybdate (lane 4), or 250 μM ADP (lane 5) in reaction buffer (15-μl reaction mixture) at 24°C for 15 min. All reactions were stopped by adding gel loading buffer. Phosphorylated proteins were separated by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide) and visualized using a phosphorimager screen.
FIG. 6.
Transcriptional regulation of the narT gene. The autoradiograph shows the reverse transcripts obtained with 0.5 pmol of IRD800-labeled _narT_-specific primer and RNA isolated from wild-type (20 μg) and mutant m1 (40 μg) cells grown aerobically (Ae) or anaerobically (An) in the presence or absence of nitrate (20 mM) or nitrite (2 mM). Lanes A, C, G, and T contained the respective sequencing reaction mixture.
FIG. 7.
Nucleotide sequence of the narGHJI, nir, and narT promoter regions. The transcriptional start sites (+1) were determined by primer extension analysis. The predicted −10 and −35 regions are underlined. Arrows indicate inverted repeats, and shaded boxes mark NreC recognition sites. The sequences of the oligonucleotides used for the band shift assays are underlined and shown in boldface. The lengths of oligonucleotides are indicated. Numbering refers to the sequence of the complete _nir_-nar locus available from GenBank under accession no. AF029224.
FIG. 8.
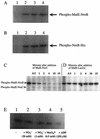
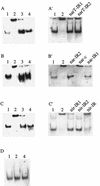
Gel mobility shift assays with purified MalE-NreC and digoxigenin-labeled DNA fragments comprising the narT (A and A′), narGHJI (B and B′), nir (C and C′), and moeB (D) promoters. Lanes: 1, promoter fragment; 2, promoter fragment plus MalE-NreC; 3, promoter fragment plus MalE-NreC plus unlabeled promoter fragment; 4, promoter fragment plus MalE-LacZα. Unlabeled oligonucleotides (A′, narT-IR1 and narT-IR2 of the narT promoter; B′, nar-IR2, nar-c, and nar-IR1 of the narGHJI promoter; and C′, nir-IR1, nir-IR2, and nir-IR of the nir promoter [locations are shown in Fig. 7]) were employed as competitive DNA. (For details, see Materials and Methods.)
FIG. 9.
Northern blot analyses of the nir operon. Total RNA isolated from wild-type (lanes 1 and 2) and mutant m1 (lanes 3 and 4) (30 μg each) cells grown anaerobically (An) with (+ nitrite) or without (−) 2 mM nitrite, separated by gel electrophoresis, was transferred to a positively charged nylon membrane and probed with a digoxigenin-labeled antisense RNA fragment comprising the entire sirA gene, approximately 300 bp of the 5′ end of nirB, and approximately 100 bp of the 3′ end of nirR. For chemiluminescent detection, CSPD (Roche) was used. The full-length transcript of the nir operon (approximately 5 kb) is indicated by an arrow. As a standard (lane 5), 90 ng of digoxigenin-labeled RNA molecular weight marker I (Roche Molecular Biochemicals) was used.
Similar articles
- Nitrate/oxygen co-sensing by an NreA/NreB sensor complex of Staphylococcus carnosus.
Nilkens S, Koch-Singenstreu M, Niemann V, Götz F, Stehle T, Unden G. Nilkens S, et al. Mol Microbiol. 2014 Jan;91(2):381-93. doi: 10.1111/mmi.12464. Epub 2013 Dec 6. Mol Microbiol. 2014. PMID: 24261791 - Control of the bifunctional O2 -sensor kinase NreB of Staphylococcus carnosus by the nitrate sensor NreA: Switching from kinase to phosphatase state.
Klein R, Kretzschmar AK, Unden G. Klein R, et al. Mol Microbiol. 2020 Feb;113(2):369-380. doi: 10.1111/mmi.14425. Epub 2019 Dec 12. Mol Microbiol. 2020. PMID: 31732993 - Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli.
Stewart V. Stewart V. Mol Microbiol. 1993 Aug;9(3):425-34. doi: 10.1111/j.1365-2958.1993.tb01704.x. Mol Microbiol. 1993. PMID: 8412692 Review. - Nitrate reductases in Escherichia coli.
Bonnefoy V, Demoss JA. Bonnefoy V, et al. Antonie Van Leeuwenhoek. 1994;66(1-3):47-56. doi: 10.1007/BF00871632. Antonie Van Leeuwenhoek. 1994. PMID: 7747940 Review.
Cited by
- Proteomics fingerprinting reveals importance of iron and oxidative stress in _Streptomyces scabies_-Solanum tuberosum interactions.
Giroux L, Isayenka I, Lerat S, Beaudoin N, Beaulieu C. Giroux L, et al. Front Microbiol. 2024 Oct 2;15:1466927. doi: 10.3389/fmicb.2024.1466927. eCollection 2024. Front Microbiol. 2024. PMID: 39417082 Free PMC article. - Bacterial iron-sulfur regulatory proteins as biological sensor-switches.
Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. Crack JC, et al. Antioxid Redox Signal. 2012 Nov 1;17(9):1215-31. doi: 10.1089/ars.2012.4511. Epub 2012 Mar 6. Antioxid Redox Signal. 2012. PMID: 22239203 Free PMC article. Review. - The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus.
Yan M, Yu C, Yang J, Ji Y. Yan M, et al. J Bacteriol. 2011 Apr;193(8):1799-805. doi: 10.1128/JB.01511-10. Epub 2011 Feb 18. J Bacteriol. 2011. PMID: 21335452 Free PMC article. - Staphylococcus aureus pathogenesis in diverse host environments.
Balasubramanian D, Harper L, Shopsin B, Torres VJ. Balasubramanian D, et al. Pathog Dis. 2017 Jan 1;75(1):ftx005. doi: 10.1093/femspd/ftx005. Pathog Dis. 2017. PMID: 28104617 Free PMC article. Review. - Rhein against Staphylococcus xylosus by interfering with respiratory metabolism and inducing oxidative stress.
Li Y, Chen W, Ma J, Huang G, Li G, He Q, Kong X, Tang L, Chen J, Ding W, Zhang Z, Ding W. Li Y, et al. Curr Res Food Sci. 2024 Mar 16;8:100718. doi: 10.1016/j.crfs.2024.100718. eCollection 2024. Curr Res Food Sci. 2024. PMID: 38545378 Free PMC article.
References
- Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. - PubMed
- Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. - PubMed
- Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous