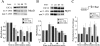Mutant DMPK 3'-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD - PubMed (original) (raw)
Mutant DMPK 3'-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD
Jeffrey D Amack et al. J Cell Biol. 2002.
Abstract
Myotonic dystrophy (DM) is caused by two similar noncoding repeat expansion mutations (DM1 and DM2). It is thought that both mutations produce pathogenic RNA molecules that accumulate in nuclear foci. The DM1 mutation is a CTG expansion in the 3' untranslated region (3'-UTR) of dystrophia myotonica protein kinase (DMPK). In a cell culture model, mutant transcripts containing a (CUG)200 DMPK 3'-UTR disrupt C2C12 myoblast differentiation; a phenotype similar to what is observed in myoblast cultures derived from DM1 patient muscle. Here, we have used our cell culture model to investigate how the mutant 3'-UTR RNA disrupts differentiation. We show that MyoD protein levels are compromised in cells that express mutant DMPK 3'-UTR transcripts. MyoD, a transcription factor required for the differentiation of myoblasts during muscle regeneration, activates differentiation-specific genes by binding E-boxes. MyoD levels are significantly reduced in myoblasts expressing the mutant 3'-UTR RNA within the first 6 h under differentiation conditions. This reduction correlates with blunted E-box-mediated gene expression at time points that are critical for initiating differentiation. Importantly, restoring MyoD levels rescues the differentiation defect. We conclude that mutant DMPK 3'-UTR transcripts disrupt myoblast differentiation by reducing MyoD levels below a threshold required to activate the differentiation program.
Figures
Figure 1.
The myogenic machinery downstream of myogenin is functional in cells expressing the mutant DMPK 3′-UTR. (A) The temporal progression of major events in the C2C12 differentiation pathway. Cells proliferating in growth media express MyoD and Myf5. When cultured in differentiation media lacking growth factors, cells initiate myogenin expression, exit the cell cycle, turn on muscle-specific structural genes, such as MHC, and fuse into myotubes. (B) Uninfected GFP+mut 3′-UTR pool (mut 3′-UTR) cells show a differentiation defect, and do not fuse into myotubes (stained red by immunofluorescent staining of MHC) as effectively as GFP+wt 3′-UTR pool (wt 3′-UTR) and control C2C12 cells. However, GFP+mut 3′-UTR pool cells infected with a retrovirus that produces myogenin are capable of forming thick myotubes similar to those formed in GFP+wt 3′-UTR pool and C2C12 cells. Infection with a control retrovirus that expresses LacZ has no effect on the differentiation phenotype. Nuclei are stained blue with DAPI. (C) Western blot analysis of cultures in growth media shows exogenous myogenin expression only in cells infected with the myogenin retrovirus (+myg). These blots were also probed for dynein, which serves as a loading control.
Figure 1.
The myogenic machinery downstream of myogenin is functional in cells expressing the mutant DMPK 3′-UTR. (A) The temporal progression of major events in the C2C12 differentiation pathway. Cells proliferating in growth media express MyoD and Myf5. When cultured in differentiation media lacking growth factors, cells initiate myogenin expression, exit the cell cycle, turn on muscle-specific structural genes, such as MHC, and fuse into myotubes. (B) Uninfected GFP+mut 3′-UTR pool (mut 3′-UTR) cells show a differentiation defect, and do not fuse into myotubes (stained red by immunofluorescent staining of MHC) as effectively as GFP+wt 3′-UTR pool (wt 3′-UTR) and control C2C12 cells. However, GFP+mut 3′-UTR pool cells infected with a retrovirus that produces myogenin are capable of forming thick myotubes similar to those formed in GFP+wt 3′-UTR pool and C2C12 cells. Infection with a control retrovirus that expresses LacZ has no effect on the differentiation phenotype. Nuclei are stained blue with DAPI. (C) Western blot analysis of cultures in growth media shows exogenous myogenin expression only in cells infected with the myogenin retrovirus (+myg). These blots were also probed for dynein, which serves as a loading control.
Figure 1.
The myogenic machinery downstream of myogenin is functional in cells expressing the mutant DMPK 3′-UTR. (A) The temporal progression of major events in the C2C12 differentiation pathway. Cells proliferating in growth media express MyoD and Myf5. When cultured in differentiation media lacking growth factors, cells initiate myogenin expression, exit the cell cycle, turn on muscle-specific structural genes, such as MHC, and fuse into myotubes. (B) Uninfected GFP+mut 3′-UTR pool (mut 3′-UTR) cells show a differentiation defect, and do not fuse into myotubes (stained red by immunofluorescent staining of MHC) as effectively as GFP+wt 3′-UTR pool (wt 3′-UTR) and control C2C12 cells. However, GFP+mut 3′-UTR pool cells infected with a retrovirus that produces myogenin are capable of forming thick myotubes similar to those formed in GFP+wt 3′-UTR pool and C2C12 cells. Infection with a control retrovirus that expresses LacZ has no effect on the differentiation phenotype. Nuclei are stained blue with DAPI. (C) Western blot analysis of cultures in growth media shows exogenous myogenin expression only in cells infected with the myogenin retrovirus (+myg). These blots were also probed for dynein, which serves as a loading control.
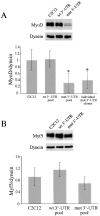
Figure 2.
mut 3′-UTR-res cells express elevated levels of MyoD. (A) An individual GFP+mut 3′-UTR clone, named mut 3′-UTR-res, expresses mutant DMPK 3′-UTR transcripts, which are detected in aggregate foci (red) by RNA FISH, but retain the ability to fuse into multinucleated myotubes (stained green by GFP expression). Nuclei are stained blue with DAPI. (B) RNA slot blot analysis shows that steady-state MyoD levels are an average 2.6-fold higher in mut 3′-UTR-res cells than in C2C12 cells (n = 3 experiments). MyoD levels were normalized to gapdh levels. (C) Western blotting revealed that MyoD protein levels are elevated in mut 3′-UTR-res cells as compared with C2C12 cells, but only by an average of 1.2-fold (n = 3 experiments). MyoD protein levels were normalized to dynein.
Figure 3.
Protein levels of MyoD, but not Myf5, are low in proliferating GFP+mut 3′-UTR cells. (A) Despite variability, Western blots indicate that MyoD levels are significantly lower in proliferating GFP+mut 3′-UTR pool (mut 3′-UTR) cells as compared with GFP+wt 3′-UTR (wt 3′-UTR) and control C2C12 cells. Western blot results from six independent experiments were quantified by densitometry. The graph shows the average MyoD levels (normalized to dynein). Also shown is the average MyoD level among six individual GFP+mut 3′-UTR clones, which is similar to the average in the GFP+mut 3′-UTR pool. An asterisk (*) signifies a statistical difference (P < 0.05) when compared with both C2C12 and GFP+wt 3′-UTR pool cells. (B) Myf5 levels are also variable in proliferating cells, but quantification of Myf5 Western blots (n = 3 experiments) showed no significant differences among C2C12, GFP+wt 3′-UTR pool, and GFP+mut 3′-UTR pool cells. Error bars represent one standard deviation.
Figure 4.
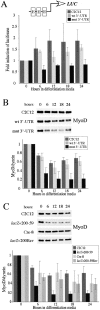
MyoD protein levels are reduced and E-box–luciferase activation is blunted in GFP+mut 3′-UTR cells after 24 h in differentiation media. (A) The pattern of MyoD expression in control C2C12, GFP+wt 3′-UTR pool, and GFP+mut 3′-UTR pool cells cultured in differentiation media was analyzed by Western blots. A representative Western blot is shown, and the graph reports the average behavior of MyoD expression in each cell population, as determined by quantifying results from five independent experiments. MyoD levels are normalized to dynein levels, which do not change during differentiation, and are analyzed relative to the levels measured in proliferating cells (0 h in differentiation media). The difference in the MyoD expression pattern between GFP+mut 3′-UTR pool cells and both C2C12 and GFP+wt 3′-UTR pool cells is statistically significant (P < 0.05) only after 24 h (*). (B) A parallel analysis of the Myf5 expression pattern revealed no statistically significant differences at any time point between the GFP+mut 3′-UTR pool and the control C2C12 and GFP+wt 3′-UTR pool populations. (C) A luciferase (LUC) reporter construct containing three E-boxes was used to measure E-box–mediated gene expression in control C2C12, GFP+wt 3′-UTR pool, and GFP+mut 3′-UTR pool cells. The graph shows the average fold induction of E-box–luciferase expression relative to luciferase levels in proliferating cells (from three independent experiments). All error bars represent one standard deviation.
Figure 5.
E-box–luciferase activation and MyoD protein levels are compromised in cells expressing the mutant DMPK 3′-UTR within the first 6 h in differentiation media. (A) The average fold induction of E-box–luciferase expression in C2C12, GFP+wt 3′-UTR pool, and GFP+mut 3′-UTR pool cells during the first 24 h in differentiation media (from three experiments). (B) Western analysis at these time points revealed a significant reduction in MyoD protein levels in the GFP+mut 3′-UTR pool as compared with control cells. The graph shows averaged results from three experiments. (C) Analysis of MyoD protein levels in lacZ+200–59 cells, which express the mutant DMPK 3′-UTR RNA, and lacZ+200–59Rev and Cre-8 cells, which have had expression of the RNA ablated, demonstrates that the reduction of MyoD is due to the presence of the mutant DMPK 3′-UTR transcript. Again, the graph shows results from three experiments. All error bars represent one standard deviation.
Figure 6.
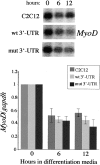
Reduced MyoD protein levels in GFP+mut 3′-UTR cells do not result from differential MyoD transcription or RNA stability. MyoD expression in control C2C12 cells, the GFP+wt 3′-UTR pool, and the GFP+mut 3′-UTR pool after 0, 6, and 12 h in differentiation media were analyzed on Northern blots and RNA slot blots. A Northern blot is shown, and the graph shows the behavior of MyoD steady-state RNA levels (normalized to gapdh levels) after 6 and 12 h in differentiation media relative to levels present in proliferating cells. These results were quantified from three slot blot experiments.
Figure 7.
Exogenous MyoD rescues the differentiation defect in cells expressing the mutant DMPK 3′-UTR RNA. (A) GFP+mut 3′-UTR pool cells infected with a retrovirus that expresses MyoD regain the ability to form myotubes (stained red by immunofluorescent staining of MHC) as effectively as infected control C2C12 and GFP+wt 3′-UTR pool cultures. Nuclei are stained blue with DAPI. (B) Western blotting shows increased expression of MyoD protein in proliferating cells infected with the MyoD retrovirus. The graph shows relative MyoD levels (normalized to dynein) in infected and uninfected cells. Similar results were seen in three separate infections. (C) Analysis of myogenin protein expression in infected and uninfected cells cultured in differentiation media shows that exogenous MyoD expression corrects the defect in myogenin up-regulation in GFP+mut 3′-UTR pool cells.
Figure 7.
Exogenous MyoD rescues the differentiation defect in cells expressing the mutant DMPK 3′-UTR RNA. (A) GFP+mut 3′-UTR pool cells infected with a retrovirus that expresses MyoD regain the ability to form myotubes (stained red by immunofluorescent staining of MHC) as effectively as infected control C2C12 and GFP+wt 3′-UTR pool cultures. Nuclei are stained blue with DAPI. (B) Western blotting shows increased expression of MyoD protein in proliferating cells infected with the MyoD retrovirus. The graph shows relative MyoD levels (normalized to dynein) in infected and uninfected cells. Similar results were seen in three separate infections. (C) Analysis of myogenin protein expression in infected and uninfected cells cultured in differentiation media shows that exogenous MyoD expression corrects the defect in myogenin up-regulation in GFP+mut 3′-UTR pool cells.
Figure 7.
Exogenous MyoD rescues the differentiation defect in cells expressing the mutant DMPK 3′-UTR RNA. (A) GFP+mut 3′-UTR pool cells infected with a retrovirus that expresses MyoD regain the ability to form myotubes (stained red by immunofluorescent staining of MHC) as effectively as infected control C2C12 and GFP+wt 3′-UTR pool cultures. Nuclei are stained blue with DAPI. (B) Western blotting shows increased expression of MyoD protein in proliferating cells infected with the MyoD retrovirus. The graph shows relative MyoD levels (normalized to dynein) in infected and uninfected cells. Similar results were seen in three separate infections. (C) Analysis of myogenin protein expression in infected and uninfected cells cultured in differentiation media shows that exogenous MyoD expression corrects the defect in myogenin up-regulation in GFP+mut 3′-UTR pool cells.
Figure 8.
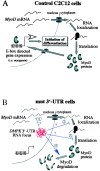
A model for how mutant DMPK 3′-UTR transcripts reduce MyoD protein levels and disrupt C2C12 differentiation. (A) A schematic of steps involved in producing MyoD in C2C12 myoblasts. MyoD, which is required for differentiation of adult myoblasts, ultimately initiates differentiation genes, such as myogenin, by binding E-boxes. (B) Potential steps where mutant DMPK 3′-UTR RNA (red knot in the nucleus) could interfere with MyoD protein production. MyoD mRNA transcription, stability, and transport are unaffected, but the mutant 3′-UTR RNA could alter splicing or processing of the MyoD transcript (1), RNA localization in the cytoplasm (2), translation efficiency (3), or protein stability (4). The end result is a reduction of MyoD protein levels, which fall below a critical threshold required to effectively activate gene expression and initiate differentiation.
Similar articles
- The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation.
Amack JD, Mahadevan MS. Amack JD, et al. Hum Mol Genet. 2001 Sep 1;10(18):1879-87. doi: 10.1093/hmg/10.18.1879. Hum Mol Genet. 2001. PMID: 11555624 - Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model.
Amack JD, Paguio AP, Mahadevan MS. Amack JD, et al. Hum Mol Genet. 1999 Oct;8(11):1975-84. doi: 10.1093/hmg/8.11.1975. Hum Mol Genet. 1999. PMID: 10484765 - Inhibition of myogenesis in transgenic mice expressing the human DMPK 3'-UTR.
Storbeck CJ, Drmanic S, Daniel K, Waring JD, Jirik FR, Parry DJ, Ahmed N, Sabourin LA, Ikeda JE, Korneluk RG. Storbeck CJ, et al. Hum Mol Genet. 2004 Mar 15;13(6):589-600. doi: 10.1093/hmg/ddh064. Epub 2004 Jan 20. Hum Mol Genet. 2004. PMID: 14734627 - Tackling the pathogenesis of RNA nuclear retention in myotonic dystrophy.
Mastroyiannopoulos NP, Shammas C, Phylactou LA. Mastroyiannopoulos NP, et al. Biol Cell. 2010 Jul 23;102(9):515-23. doi: 10.1042/BC20100040. Biol Cell. 2010. PMID: 20690904 Review. - Myotonic dystrophy: emerging mechanisms for DM1 and DM2.
Cho DH, Tapscott SJ. Cho DH, et al. Biochim Biophys Acta. 2007 Feb;1772(2):195-204. doi: 10.1016/j.bbadis.2006.05.013. Epub 2006 Jun 20. Biochim Biophys Acta. 2007. PMID: 16876389 Review.
Cited by
- Pluripotent Stem Cells in Disease Modeling and Drug Discovery for Myotonic Dystrophy Type 1.
Bérenger-Currias N, Martinat C, Baghdoyan S. Bérenger-Currias N, et al. Cells. 2023 Feb 10;12(4):571. doi: 10.3390/cells12040571. Cells. 2023. PMID: 36831237 Free PMC article. Review. - Inhibition of Postn Rescues Myogenesis Defects in Myotonic Dystrophy Type 1 Myoblast Model.
Shen X, Liu Z, Wang C, Xu F, Zhang J, Li M, Lei Y, Wang A, Bi C, Zhu G. Shen X, et al. Front Cell Dev Biol. 2021 Aug 19;9:710112. doi: 10.3389/fcell.2021.710112. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34490258 Free PMC article. - RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression.
Yadava RS, Frenzel-McCardell CD, Yu Q, Srinivasan V, Tucker AL, Puymirat J, Thornton CA, Prall OW, Harvey RP, Mahadevan MS. Yadava RS, et al. Nat Genet. 2008 Jan;40(1):61-8. doi: 10.1038/ng.2007.28. Epub 2007 Dec 16. Nat Genet. 2008. PMID: 18084293 Free PMC article. - Modeling muscle regeneration in RNA toxicity mice.
Yadava RS, Mandal M, Giese JM, Rigo F, Bennett CF, Mahadevan MS. Yadava RS, et al. Hum Mol Genet. 2021 Jun 9;30(12):1111-1130. doi: 10.1093/hmg/ddab108. Hum Mol Genet. 2021. PMID: 33864373 Free PMC article. - miR-322/-503 rescues myoblast defects in myotonic dystrophy type 1 cell model by targeting CUG repeats.
Shen X, Xu F, Li M, Wu S, Zhang J, Wang A, Xu L, Liu Y, Zhu G. Shen X, et al. Cell Death Dis. 2020 Oct 22;11(10):891. doi: 10.1038/s41419-020-03112-6. Cell Death Dis. 2020. PMID: 33093470 Free PMC article.
References
- Amack, J.D., and M.S. Mahadevan. 2001. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 10:1879–1887. - PubMed
- Amack, J.D., A.P. Paguio, and M.S. Mahadevan. 1999. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum. Mol. Genet. 8:1975–1984. - PubMed
- Bergstrom, D.A., B.H. Penn, A. Strand, R.L. Perry, M.A. Rudnicki, and S.J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 9:587–600. - PubMed
- Blau, H.M., C.P. Chiu, and C. Webster. 1983. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 32:1171–1180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases