A novel role for p120 catenin in E-cadherin function - PubMed (original) (raw)
. 2002 Nov 11;159(3):465-76.
doi: 10.1083/jcb.200205115. Epub 2002 Nov 11.
Michael A Davis, Jolanda van Hengel, Deborah J Mariner, Kirk Barnes, Molly A Thoreson, Panos Z Anastasiadis, Linsey Matrisian, Linda M Bundy, Linda Sealy, Barbara Gilbert, Frans van Roy, Albert B Reynolds
Affiliations
- PMID: 12427869
- PMCID: PMC2173073
- DOI: 10.1083/jcb.200205115
A novel role for p120 catenin in E-cadherin function
Renee C Ireton et al. J Cell Biol. 2002.
Abstract
Indirect evidence suggests that p120-catenin (p120) can both positively and negatively affect cadherin adhesiveness. Here we show that the p120 gene is mutated in SW48 cells, and that the cadherin adhesion system is impaired as a direct consequence of p120 insufficiency. Restoring normal levels of p120 caused a striking reversion from poorly differentiated to cobblestone-like epithelial morphology, indicating a crucial role for p120 in reactivation of E-cadherin function. The rescue efficiency was enhanced by increased levels of p120, and reduced by the presence of the phosphorylation domain, a region previously postulated to confer negative regulation. Surprisingly, the rescue was associated with substantially increased levels of E-cadherin. E-cadherin mRNA levels were unaffected by p120 expression, but E-cadherin half-life was more than doubled. Direct p120-E-cadherin interaction was crucial, as p120 deletion analysis revealed a perfect correlation between E-cadherin binding and rescue of epithelial morphology. Interestingly, the epithelial morphology could also be rescued by forced expression of either WT E-cadherin or a p120-uncoupled mutant. Thus, the effects of uncoupling p120 from E-cadherin can be at least partially overcome by artificially maintaining high levels of cadherin expression. These data reveal a cooperative interaction between p120 and E-cadherin and a novel role for p120 that is likely indispensable in normal cells.
Figures
Figure 1.
Characterization of the E-cadherin complex in SW48 carcinoma cells. (A) Members of the cadherin complex were localized in SW48 cells by immunofluorescence. SW48 cells organize aberrantly into linear arrays and do not form compact epithelial colonies. p120 was localized with mAb pp120, which binds a COOH-terminal epitope present in all known p120 isoforms. (B) Cadherin complex proteins were analyzed by Western blotting of NP-40 cell lysates derived from the colon carcinoma cell lines HCT116, HCA7, and SW48. p120 is not detected in SW48 cells by mAb pp120, indicating that the COOH terminus is absent.
Figure 2.
Characterization of mutated p120 alleles in SW48 cells. (A) Schematic diagram of human p120 protein and cDNA structure. N1–N4 are the four alternatively used start codons. A, B, C, and D, are alternatively spliced exons. Individual ARM repeats are numbered. Note the phosphorylation domain between N3 and N4, immediately NH2-terminal to the ARM domain. Approximate epitopes of mAbs are indicated. (B) Primer sets used for RT-PCR and genomic DNA analysis. (C) A nonsense mutation CGA to TGA in exon 7 is indicated by the C to T mutation detected in genomic DNA and cDNA clones. The normal ATG and TAG codons are indicated. (D) Aberrant mRNA forms detected by RT-PCR. The exon structure of p120 cDNA COOH-terminal end is shown across the top. Aberrant mRNAs, each resulting in premature stop codons, are located below. Aberrant splicing in the 3′-terminal part of p120 mRNA results in the indicated gene products, which include retention of intron 19, retention of both introns 19 and 20, or exclusion of exon 17.
Figure 3.
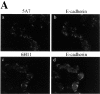
Antibodies against the NH2 terminus of p120 detect extremely low levels of p120 in SW48 cells. (A) p120 localization with NH2-terminally directed p120 mAbs. SW48 cells were costained with p120 mAb's 5A7 or 6H11 (a and c), and E-cadherin mAb C-20820 (b and d). (B) Quantitative analysis of p120 expression was performed by immunoprecipitating p120 from RIPA cell lysates with either NH2-terminally directed mAbs 5A7 or 6H11, or the COOH-terminally directed mAb pp120 (top). The immunoprecipitates were split and Western blotted with mAb 5A7 (lanes 1–6) or mAb pp120 (lanes 7–12). HCA7 cells expressing normal levels of p120 were used for comparison. The blots are overexposed to visualize the extreme low levels of p120.
Figure 3.
Antibodies against the NH2 terminus of p120 detect extremely low levels of p120 in SW48 cells. (A) p120 localization with NH2-terminally directed p120 mAbs. SW48 cells were costained with p120 mAb's 5A7 or 6H11 (a and c), and E-cadherin mAb C-20820 (b and d). (B) Quantitative analysis of p120 expression was performed by immunoprecipitating p120 from RIPA cell lysates with either NH2-terminally directed mAbs 5A7 or 6H11, or the COOH-terminally directed mAb pp120 (top). The immunoprecipitates were split and Western blotted with mAb 5A7 (lanes 1–6) or mAb pp120 (lanes 7–12). HCA7 cells expressing normal levels of p120 were used for comparison. The blots are overexposed to visualize the extreme low levels of p120.
Figure 4.
p120 expression in SW48 cells rescues epithelial colony morphology. (A and B) SW48 cells were infected with either LZRS- MS-IRES-GFP (LZRS; a, b, e, and f), or LZRS-p120-IRES-GFP (LZRS-p120; c, d, g, and h). Infected cells were sparsely plated and cultured until individual colonies appeared. (A) Rescue of epithelial morphology by p120. To visualize GFP, cells were fixed with paraformaldehyde to preserve the GFP signal (a–d). p120 was visualized with mAb 15D2 (a and c) and GFP with direct UV microscopy (b and d). GFP expression alone (b) had no effect on SW48 cell morphology. (B) Colocalization of p120 and E-cadherin in infected SW48 cells. To use the green wavelength for E-cadherin staining, cells were fixed in methanol to eliminate the GFP signal and then costained for p120 (e and g) and E-cadherin (f and h). (C) Organization of the LZRS-p120-GFP retroviral vector. An IRES is sandwiched between p120 and GFP cDNAs to allow for translation of both genes from a single mRNA transcript. Thus, the levels of GFP and p120 in infected cells are linked. Key functional elements of the vector are indicated.
Figure 4.
p120 expression in SW48 cells rescues epithelial colony morphology. (A and B) SW48 cells were infected with either LZRS- MS-IRES-GFP (LZRS; a, b, e, and f), or LZRS-p120-IRES-GFP (LZRS-p120; c, d, g, and h). Infected cells were sparsely plated and cultured until individual colonies appeared. (A) Rescue of epithelial morphology by p120. To visualize GFP, cells were fixed with paraformaldehyde to preserve the GFP signal (a–d). p120 was visualized with mAb 15D2 (a and c) and GFP with direct UV microscopy (b and d). GFP expression alone (b) had no effect on SW48 cell morphology. (B) Colocalization of p120 and E-cadherin in infected SW48 cells. To use the green wavelength for E-cadherin staining, cells were fixed in methanol to eliminate the GFP signal and then costained for p120 (e and g) and E-cadherin (f and h). (C) Organization of the LZRS-p120-GFP retroviral vector. An IRES is sandwiched between p120 and GFP cDNAs to allow for translation of both genes from a single mRNA transcript. Thus, the levels of GFP and p120 in infected cells are linked. Key functional elements of the vector are indicated.
Figure 5.
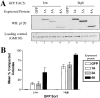
Epithelial rescue by p120 is both expression level and isoform dependent. (A) Isolation by GFP FACS of cell populations expressing low or high levels of p120 isoforms. Cells were infected with viruses containing GFP alone (lanes 1 and 5) or GFP linked by an IRES to p120 isoforms 1A (lanes 2 and 6), 3A (lanes 3 and 7), or 4A (lanes 4 and 8). Cells were sorted by FACS for low (lanes 1–4) and high (lanes 5–8) GFP expression. Protein normalization was confirmed by Western blotting with GM130 antibody. Note the tight correlation between GFP and p120 levels. Each isoform expression level is roughly equivalent within low and high level groups. (B) Quantification of the potency of p120 rescue as a function of expression level and isoform. Cells sorted as described above were plated at low density, grown into clonal colonies, and the colonies scored as compact or loose. The data reflect the ratio of compacted (rescued) colonies divided by the total number of colonies. Compacted colonies occurred more frequently when p120 is expressed at high levels. p120ctn4, which lacks the NH2-terminal phosphorylation domain, is more efficient at inducing compaction than p120ctn1 and 3, which retain this domain.
Figure 6.
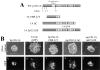
The p120 COOH terminus is dispensable for epithelial rescue. (A) Schematic of WT-p120 and p120 variants representing the mutants found in SW48 cells. Alternative ATG start sites are indicated. Light gray boxes represent ARM repeats. mp120–1A Δ622–628 contains a six-aa deletion in ARM 6 postulated to uncouple p120 from RhoA. (B) Effects of overexpression of p120 mutants in SW48 cells. Mutant constructs were cloned into pLZRS-p120-IRES-GFP and expressed in SW48 cells by retroviral infection. Infected cells were collected by gated GFP FACS, plated at low density, and colonies were photographed 1 wk later. Top panels are phase contrast images of colonies and bottom panels show GFP fluorescent images of the same cells. hp120–3A 1908 C/T does not affect the morphology (e and f), but both mp120–1A ΔC and mp120–1A Δ622–628 efficiently induce compaction (g–j).
Figure 7.
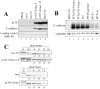
p120-induced compaction is dependent on direct interaction between p120 and E-cadherin. (A) Structure-function analysis of p120 sequences necessary for interaction with E-cadherin. Each of the ten p120 ARM repeats was individually deleted, expressed in SW48 cells, and polyclonal cell lines derived by GFP FACS. E-cadherin immunoprecipitates were isolated, divided in half, and Western blotted for E-cadherin (middle) or p120 (top). p120 immunoprecipitation and Western blot with mAb p120 controlled for the exogenous expression of p120 mutants (bottom). (*) denotes p120 degradation products. Controls include cells expressing WT-p120 (lane 12), cells infected with empty vector (lane 11), and cells immunoprecipitated with an irrelevant nonspecific antibody (NS). (B) Effects of above constructs in SW48 cells assayed by immunofluorescence for p120 (top) and E-cadherin (bottom). Deletion of ARM repeats 1–5 and 7 block rescue of compaction. ARM repeats 6 and 8–10 are dispensable. E-cadherin binding (A) correlates perfectly with the ability to rescue epithelial morphology (B).
Figure 7.
p120-induced compaction is dependent on direct interaction between p120 and E-cadherin. (A) Structure-function analysis of p120 sequences necessary for interaction with E-cadherin. Each of the ten p120 ARM repeats was individually deleted, expressed in SW48 cells, and polyclonal cell lines derived by GFP FACS. E-cadherin immunoprecipitates were isolated, divided in half, and Western blotted for E-cadherin (middle) or p120 (top). p120 immunoprecipitation and Western blot with mAb p120 controlled for the exogenous expression of p120 mutants (bottom). (*) denotes p120 degradation products. Controls include cells expressing WT-p120 (lane 12), cells infected with empty vector (lane 11), and cells immunoprecipitated with an irrelevant nonspecific antibody (NS). (B) Effects of above constructs in SW48 cells assayed by immunofluorescence for p120 (top) and E-cadherin (bottom). Deletion of ARM repeats 1–5 and 7 block rescue of compaction. ARM repeats 6 and 8–10 are dispensable. E-cadherin binding (A) correlates perfectly with the ability to rescue epithelial morphology (B).
Figure 8.
p120 expression stabilizes E-cadherin by a post-transcriptional mechanism. (A) SW48 cells were infected with LZRS-p120-IRES-neo, and stable cell lines isolated. Levels of E-cadherin were assayed by Western blotting RIPA cell lysates. Parental (lane 1) or cell lines expressing neo only (lanes 2 and 3) were compared with two separately derived p120 expressing cell lines (lanes 4 and 5) and to HCT116 cells (which express normal levels of p120 and E-cadherin). GM130 blotting (bottom) was used as a loading control. p120 expression increased E-cadherin levels by more than fivefold in SW48 cells. (B) E-cadherin mRNA levels are unchanged by p120 expression. Total RNA was isolated and compared by Northern analysis using a human E-cadherin cDNA probe. To control for loading, the blot was stripped and reprobed with GAPDH cDNA. MDA-MB-231 cells (lane 1) do not express E-cadherin mRNA. SW48 cells stably expressing p120–4A (lane 2), p120–3A (lane 3), or neo alone (lane 4), are compared with SW48 parental (lane 5), HCA7 (lane 6), and HCT116 cells (lane 7). (C) Analysis of E-cadherin stability in p120-expressing cells. Clonal (p120 3A/neo-15) or polyclonal (p120–3A/neo) cell lines were generated by infection. E-cadherin half life was ascertained by pulse-chase analysis and compared with control clonal (neo-18) and polyclonal (neo) cell lines. Chase times are indicated across the top. Densitometric analyses are located below each lane and represent a normalized value where the 4 h chase lane (top) or the 2 h chase lane (bottom) represent the value 1.0. p120 expression essentially doubles the half life of E-cadherin.
Figure 9.
Epithelial rescue by overexpression of WT or p120-uncoupled E-cadherin in SW48 cells. SW48 cells were infected with LZRS-neo viruses containing p120, E-cadherin, or p120-uncoupled E-cadherin (E-cadherin-764), and colonies generated by G418 selection. Cells are stained for immunofluorescence using E-cadherin mAb C-20820. Expression of neo alone (a) had no effect on the parental SW48 phenotype (b). However, WT-E-cadherin (panel d) and p120-uncoupled E-cadherin (e) induced an epithelial morphology similar to that induced by p120 expression (c).
Similar articles
- Regulation of p120-catenin nucleocytoplasmic shuttling activity.
Roczniak-Ferguson A, Reynolds AB. Roczniak-Ferguson A, et al. J Cell Sci. 2003 Oct 15;116(Pt 20):4201-12. doi: 10.1242/jcs.00724. Epub 2003 Sep 2. J Cell Sci. 2003. PMID: 12953069 - The regulatory or phosphorylation domain of p120 catenin controls E-cadherin dynamics at the plasma membrane.
Fukumoto Y, Shintani Y, Reynolds AB, Johnson KR, Wheelock MJ. Fukumoto Y, et al. Exp Cell Res. 2008 Jan 1;314(1):52-67. doi: 10.1016/j.yexcr.2007.07.024. Epub 2007 Jul 31. Exp Cell Res. 2008. PMID: 17719574 Free PMC article. - Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion.
Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. Thoreson MA, et al. J Cell Biol. 2000 Jan 10;148(1):189-202. doi: 10.1083/jcb.148.1.189. J Cell Biol. 2000. PMID: 10629228 Free PMC article. - Emerging roles for p120-catenin in cell adhesion and cancer.
Reynolds AB, Roczniak-Ferguson A. Reynolds AB, et al. Oncogene. 2004 Oct 18;23(48):7947-56. doi: 10.1038/sj.onc.1208161. Oncogene. 2004. PMID: 15489912 Review. - Protecting your tail: regulation of cadherin degradation by p120-catenin.
Kowalczyk AP, Reynolds AB. Kowalczyk AP, et al. Curr Opin Cell Biol. 2004 Oct;16(5):522-7. doi: 10.1016/j.ceb.2004.07.001. Curr Opin Cell Biol. 2004. PMID: 15363802 Review.
Cited by
- Cell surface localization of α3β4 nicotinic acetylcholine receptors is regulated by N-cadherin homotypic binding and actomyosin contractility.
Brusés JL. Brusés JL. PLoS One. 2013 Apr 23;8(4):e62435. doi: 10.1371/journal.pone.0062435. Print 2013. PLoS One. 2013. PMID: 23626818 Free PMC article. - Galectin-3 protein regulates mobility of N-cadherin and GM1 ganglioside at cell-cell junctions of mammary carcinoma cells.
Boscher C, Zheng YZ, Lakshminarayan R, Johannes L, Dennis JW, Foster LJ, Nabi IR. Boscher C, et al. J Biol Chem. 2012 Sep 21;287(39):32940-52. doi: 10.1074/jbc.M112.353334. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22846995 Free PMC article. - Neural Wiskott-Aldrich syndrome protein (N-WASP)-mediated p120-catenin interaction with Arp2-Actin complex stabilizes endothelial adherens junctions.
Rajput C, Kini V, Smith M, Yazbeck P, Chavez A, Schmidt T, Zhang W, Knezevic N, Komarova Y, Mehta D. Rajput C, et al. J Biol Chem. 2013 Feb 8;288(6):4241-50. doi: 10.1074/jbc.M112.440396. Epub 2012 Dec 4. J Biol Chem. 2013. PMID: 23212915 Free PMC article. - Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1.
Markham NO, Cooper T, Goff M, Gribben EM, Carnahan RH, Reynolds AB. Markham NO, et al. Hybridoma (Larchmt). 2012 Aug;31(4):246-54. doi: 10.1089/hyb.2012.0009. Hybridoma (Larchmt). 2012. PMID: 22894777 Free PMC article. - p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin.
Xiao K, Garner J, Buckley KM, Vincent PA, Chiasson CM, Dejana E, Faundez V, Kowalczyk AP. Xiao K, et al. Mol Biol Cell. 2005 Nov;16(11):5141-51. doi: 10.1091/mbc.e05-05-0440. Epub 2005 Aug 24. Mol Biol Cell. 2005. PMID: 16120645 Free PMC article.
References
- Aberle, H., S. Butz, J. Stappert, H. Weissig, R. Kemler, and H. Hoschuetzky. 1994. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 107:3655–3663. - PubMed
- Anastasiadis, P.Z., and A.B. Reynolds. 2000. The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci. 113:1319–1334. - PubMed
- Anastasiadis, P.Z., and A.B. Reynolds. 2001. Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 13:604–610. - PubMed
- Anastasiadis, P.Z., S.Y. Moon, M.A. Thoreson, D.J. Mariner, H.C. Crawford, Y. Zheng, and A.B. Reynolds. 2000. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2:637–644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous