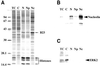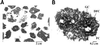Functional proteomic analysis of human nucleolus - PubMed (original) (raw)
Functional proteomic analysis of human nucleolus
Alexander Scherl et al. Mol Biol Cell. 2002 Nov.
Abstract
The notion of a "plurifunctional" nucleolus is now well established. However, molecular mechanisms underlying the biological processes occurring within this nuclear domain remain only partially understood. As a first step in elucidating these mechanisms we have carried out a proteomic analysis to draw up a list of proteins present within nucleoli of HeLa cells. This analysis allowed the identification of 213 different nucleolar proteins. This catalog complements that of the 271 proteins obtained recently by others, giving a total of approximately 350 different nucleolar proteins. Functional classification of these proteins allowed outlining several biological processes taking place within nucleoli. Bioinformatic analyses permitted the assignment of hypothetical functions for 43 proteins for which no functional information is available. Notably, a role in ribosome biogenesis was proposed for 31 proteins. More generally, this functional classification reinforces the plurifunctional nature of nucleoli and provides convincing evidence that nucleoli may play a central role in the control of gene expression. Finally, this analysis supports the recent demonstration of a coupling of transcription and translation in higher eukaryotes.
Figures
Figure 1
Analysis of the cellular fractions obtained during nucleoli purification. (A) 1-DE separation of proteins extracted from total cells (TC) and from subcellular fractions indicated on the top of the gel (C, cytoplasm; N, nuclei; Np, nucleoplasm; Nc, nucleoli). For each fraction, 10 μg protein was separated on a 12.5% polyacrylamide gel. Proteins were stained with Coomassie brilliant blue R250. Positions of B23 and histones are indicated by arrows on the right of the panel. Sizes of the molecular weight markers are indicated in kilodaltons on the left of the panel. (B, C) Western blot analyses of the different cellular fractions described in A using an anti-ERK2 antibody (B) and an anti-nucleolin antibody (C). The position of ERK2 and nucleolin is indicated by an arrow on the right of panels B and C, respectively.
Figure 2
Transmission electron micrographs of purified nucleoli. The nucleolar fraction was obtained from HeLa cells, as described in MATERIALS AND METHODS and submitted to electron microscopy analyses (A) Nucleoloar fraction at 4000× magnification. Nucleoli are the main structures observed in the last fraction of the cellular fractionation procedure. (B) Nucleolar fraction at 20,000× magnification. Purified nucleoli have conserved their characteristic ultrastructure in three main compartments: FC, fibrillar center; DFC, dense fibrillar component; and GC, granular component.
Figure 3
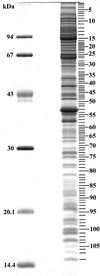
Sequential analysis of nucleolar proteins separated by SDS-PAGE. Nucleolar proteins (15 μg) were separated by SDS-PAGE on a 12.5% polyacrylamide gel and stained with Bio-Safe coomassie blue. This gel was then sequentially cut into 108 fragments. Each cut was numbered. Their position within the gel and their number are indicated by dashes on the right side of the gel. Molecular masses of known proteins separated in the same gel are indicated on the left of the figure.
Figure 4
Annotated 2-DE map of acidic nucleolar proteins. Nucleoli from HeLa cells were purified as described in MATERIALS AND METHODS. Nucleolar proteins were extracted with acetic acid before separation by 2-DE. Proteins were separated by IEF on immobilized pH gradients 4–7 in the first dimension. They were then separated by SDS-PAGE in the second dimension. Finally, proteins were identified by mass spectrometry. The image presented here is a representative gel stained with silver nitrate. The 35 identified proteins are labeled with their SWISS-PROT or TrEMBL accession numbers.
Figure 5
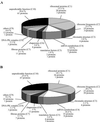
Functional classes for nucleolar proteins identified in the two independent proteomic analyses of purified human nucleoli. Functional classes were deduced for the 213 nucleolar proteins identified in this study and listed in the supplemental table, online. (A) and for 262 nucleolar proteins identified in the study of Andersen et al. (2002) (B). The name of the class with its corresponding abbreviation used in the supplemental table (C1–C10) is given. The percentage of total proteins found within each class is indicated. In addition, for each class, the number of proteins with a demonstrated involvement in the biological process is given, followed by the number of proteins with hypothetical involvement in this biological process in italics.
Similar articles
- Bioinformatic analysis of the nucleolus.
Leung AK, Andersen JS, Mann M, Lamond AI. Leung AK, et al. Biochem J. 2003 Dec 15;376(Pt 3):553-69. doi: 10.1042/BJ20031169. Biochem J. 2003. PMID: 14531731 Free PMC article. Review. - Deciphering the human nucleolar proteome.
Couté Y, Burgess JA, Diaz JJ, Chichester C, Lisacek F, Greco A, Sanchez JC. Couté Y, et al. Mass Spectrom Rev. 2006 Mar-Apr;25(2):215-34. doi: 10.1002/mas.20067. Mass Spectrom Rev. 2006. PMID: 16211575 Review. - Directed proteomic analysis of the human nucleolus.
Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Andersen JS, et al. Curr Biol. 2002 Jan 8;12(1):1-11. doi: 10.1016/s0960-9822(01)00650-9. Curr Biol. 2002. PMID: 11790298 - Proteomic profiling of the human T-cell nucleolus.
Jarboui MA, Wynne K, Elia G, Hall WW, Gautier VW. Jarboui MA, et al. Mol Immunol. 2011 Dec;49(3):441-52. doi: 10.1016/j.molimm.2011.09.005. Epub 2011 Oct 19. Mol Immunol. 2011. PMID: 22014684 - Arabidopsis nucleolar protein database (AtNoPDB).
Brown JW, Shaw PJ, Shaw P, Marshall DF. Brown JW, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D633-6. doi: 10.1093/nar/gki052. Nucleic Acids Res. 2005. PMID: 15608277 Free PMC article.
Cited by
- Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition.
Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Shav-Tal Y, et al. Mol Biol Cell. 2005 May;16(5):2395-413. doi: 10.1091/mbc.e04-11-0992. Epub 2005 Mar 9. Mol Biol Cell. 2005. PMID: 15758027 Free PMC article. - Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication.
Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. Ozawa M, et al. J Virol. 2007 Jan;81(1):30-41. doi: 10.1128/JVI.01434-06. Epub 2006 Oct 18. J Virol. 2007. PMID: 17050598 Free PMC article. - Exploring DNA-binding proteins with in vivo chemical cross-linking and mass spectrometry.
Qin H, Wang Y. Qin H, et al. J Proteome Res. 2009 Apr;8(4):1983-91. doi: 10.1021/pr8009319. J Proteome Res. 2009. PMID: 19714816 Free PMC article. - Externalized Keratin 8: A Target at the Interface of Microenvironment and Intracellular Signaling in Colorectal Cancer Cells.
Albaret MA, Vermot-Desroches C, Paré A, Roca-Martinez JX, Malet L, Esseily J, Gerossier L, Brière J, Pion N, Marcel V, Catez F, De Souza G, Vuillermoz B, Doerflinger F, Lavocat E, Subiger O, Rousset C, Bresson C, Mandon E, Jawhari A, Falson P, Jasmin M, Coute Y, Mertani HC, Saintigny P, Diaz JJ. Albaret MA, et al. Cancers (Basel). 2018 Nov 16;10(11):452. doi: 10.3390/cancers10110452. Cancers (Basel). 2018. PMID: 30453567 Free PMC article. - Subcellular localization of CIAPIN1.
Hao Z, Li X, Qiao T, Du R, Zhang G, Fan D. Hao Z, et al. J Histochem Cytochem. 2006 Dec;54(12):1437-44. doi: 10.1369/jhc.6A6960.2006. Epub 2006 Sep 6. J Histochem Cytochem. 2006. PMID: 16957168 Free PMC article.
References
- Allende JE, Allende CC. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. - PubMed
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. - PubMed
- Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. - PubMed
- Bergquist J, Gobom J, Blomberg A, Roepstorff P, Ekman R. Identification of nuclei associated proteins by 2D-gel electrophoresis and mass spectrometry. J Neurosci Methods. 2001;109:3–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials