Structure of factor-inhibiting hypoxia-inducible factor 1: An asparaginyl hydroxylase involved in the hypoxic response pathway - PubMed (original) (raw)
Structure of factor-inhibiting hypoxia-inducible factor 1: An asparaginyl hydroxylase involved in the hypoxic response pathway
Charles E Dann 3rd et al. Proc Natl Acad Sci U S A. 2002.
Abstract
Precise regulation of the evolutionarily conserved hypoxia-inducible transcription factor (HIF) ensures proper adaptation to variations in oxygen availability throughout development and into adulthood. Oxygen-dependent regulation of HIF stability and activity are mediated by hydroxylation of conserved proline and asparagine residues, respectively. Because the relevant prolyl and asparginyl hydroxylases use O(2) to effect these posttranslational modifications, these enzymes are implicated as direct oxygen sensors in the mammalian hypoxic response pathway. Here we present the structure of factor-inhibiting HIF-1 (FIH-1), the pertinent asparaginyl hydroxylase involved in hypoxic signaling. Hydroxylation of the C-terminal transactivation domain (CTAD) of HIF by FIH-1 prevents CTAD association with transcriptional coactivators under normoxic conditions. Consistent with other structurally known hydroxylases, FIH-1 is comprised of a beta-strand jellyroll core with both Fe(II) and the cosubstrate 2-oxoglutarate bound in the active site. Details of the molecular contacts at the active site of FIH-1 have been elucidated and provide a platform for future drug design. Furthermore, the structure reveals the presence of a FIH-1 homodimer that forms in solution and is essential for FIH activity.
Figures
Fig 1.
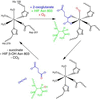
FIH-1 hydroxylates HIF-1α at Asn-803. The FIH-1 active site contains Fe(II) coordinated by three protein side chains: His-199, Asp-201, and His-279. The enzyme binds 2-OG, HIF peptide substrate, and molecular oxygen to facilitate hydroxylation of the β-carbon on Asn-803 of HIF-1α. In the course of the reaction, molecular oxygen is consumed and 2-OG is converted into succinate and carbon dioxide.
Fig 2.
The primary structure of FIH-1 is labeled with secondary structure elements taken from the x-ray crystallographic model. β-strands and helices are depicted as red arrows and yellow boxes, respectively. Residues responsible for Fe(II) binding are highlighted in red, whereas residues in close contact with 2-OG are highlighted in green.
Fig 3.
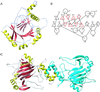
FIH-1 structure contains a β-jellyroll core marked by an extension of one of the β-sheets away from the core and helices dotting the periphery. (A) A ribbon model of the FIH-1 monomer is positioned looking between the β-sheets comprising the jellyroll and into the active site cavity. The active site metal is shown as a red sphere. Structural elements are colored as in Fig. 2. (B) A secondary structure topology diagram shows the arrangement of the 14 β-strands (triangles) and 8 helices (circles) in FIH-1. The core jellyroll motif, structurally homologous to the cupin protein family, is colored in red. (C) FIH-1 exists as a functionally relevant dimer in the crystal. The first monomer of the dimer is colored as in A, whereas the second monomer is blue. N and C termini are marked as black circles. The figure was generated by using RIBBONS (41).
Fig 4.
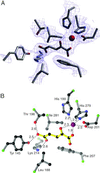
Multiple contacts are present in the FIH-1 active site. (A) A 2_F_o − _F_c simulated annealing omit map contoured at 1.1 σ reveals the presence of 2-OG in the active site. (B) 2-OG makes several hydrophilic contacts with residues of FIH-1, most notably hydrogen bonds to Lys-214 and Thr-196. Additional contacts include coordination to the Fe (red sphere) and hydrophobic interactions with Leu-188, Phe-207, and Ile-281. Ball-and-stick model in B is colored with 2-OG in yellow, Cα positions in green, carbon in black, nitrogen in blue, and oxygen in red. The figure was generated by using SETOR (42) and RIBBONS (41) modeling programs.
Fig 5.
The C terminus of FIH-1 is required for activity. (A) In vitro hydroxylation of HIF-2α 774–874 by wild-type FIH-1 inhibits interaction with p300. 35S-labeled HIF-2α 774–874 was incubated in the absence (lane 1) or presence of various recombinant MBP-FIH-1 enzymes (lanes 2–6) followed by incubation with immobilized GST-p300 CH1. 35S-labeled HIF-2α 774–874 bound to the GST-p300 CH1 domain was visualized by phosphorimaging after SDS/PAGE. Mutations that interfere with Fe(II) binding (D201A) or delete residues 303–349 compromise FIH-1 ability to block p300 association with the HIF-2α CTAD. (B) Deletion of FIH-1 residues 303–349 prevents interaction with the CTAD. 35S-labeled HIF-2α 774–874 bound to immobilized wild-type or truncated (1–302) MBP-FIH-1 was visualized after SDS/PAGE. The right lane indicates 10% of the input 35S-labeled protein in the pull-down experiments.
Similar articles
- Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases.
Koivunen P, Hirsilä M, Günzler V, Kivirikko KI, Myllyharju J. Koivunen P, et al. J Biol Chem. 2004 Mar 12;279(11):9899-904. doi: 10.1074/jbc.M312254200. Epub 2003 Dec 29. J Biol Chem. 2004. PMID: 14701857 - Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family.
Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, Ratcliffe PJ, Pugh CW, Schofield CJ. Hewitson KS, et al. J Biol Chem. 2002 Jul 19;277(29):26351-5. doi: 10.1074/jbc.C200273200. Epub 2002 May 31. J Biol Chem. 2002. PMID: 12042299 - Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
Lee C, Kim SJ, Jeong DG, Lee SM, Ryu SE. Lee C, et al. J Biol Chem. 2003 Feb 28;278(9):7558-63. doi: 10.1074/jbc.M210385200. Epub 2002 Dec 12. J Biol Chem. 2003. PMID: 12482756 - Inhibition of the Oxygen-Sensing Asparaginyl Hydroxylase Factor Inhibiting Hypoxia-Inducible Factor: A Potential Hypoxia Response Modulating Strategy.
Wu Y, Li Z, McDonough MA, Schofield CJ, Zhang X. Wu Y, et al. J Med Chem. 2021 Jun 10;64(11):7189-7209. doi: 10.1021/acs.jmedchem.1c00415. Epub 2021 May 24. J Med Chem. 2021. PMID: 34029087 Review. - Factor inhibiting hypoxia-inducible factor (FIH) and other asparaginyl hydroxylases.
Lancaster DE, McDonough MA, Schofield CJ. Lancaster DE, et al. Biochem Soc Trans. 2004 Dec;32(Pt 6):943-5. doi: 10.1042/BST0320943. Biochem Soc Trans. 2004. PMID: 15506931 Review.
Cited by
- Protein Flexibility of the α-Ketoglutarate-Dependent Oxygenase Factor-Inhibiting HIF-1: Implications for Substrate Binding, Catalysis, and Regulation.
Martin CB, Chaplin VD, Eyles SJ, Knapp MJ. Martin CB, et al. Biochemistry. 2019 Oct 1;58(39):4047-4057. doi: 10.1021/acs.biochem.9b00619. Epub 2019 Sep 20. Biochemistry. 2019. PMID: 31499004 Free PMC article. - Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification.
Kato M, Araiso Y, Noma A, Nagao A, Suzuki T, Ishitani R, Nureki O. Kato M, et al. Nucleic Acids Res. 2011 Mar;39(4):1576-85. doi: 10.1093/nar/gkq919. Epub 2010 Oct 23. Nucleic Acids Res. 2011. PMID: 20972222 Free PMC article. - Protein and DNA modifications: evolutionary imprints of bacterial biochemical diversification and geochemistry on the provenance of eukaryotic epigenetics.
Aravind L, Burroughs AM, Zhang D, Iyer LM. Aravind L, et al. Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016063. doi: 10.1101/cshperspect.a016063. Cold Spring Harb Perspect Biol. 2014. PMID: 24984775 Free PMC article. Review. - Mina, an Il4 repressor, controls T helper type 2 bias.
Okamoto M, Van Stry M, Chung L, Koyanagi M, Sun X, Suzuki Y, Ohara O, Kitamura H, Hijikata A, Kubo M, Bix M. Okamoto M, et al. Nat Immunol. 2009 Aug;10(8):872-9. doi: 10.1038/ni.1747. Epub 2009 Jun 28. Nat Immunol. 2009. PMID: 19561615 Free PMC article. - Catalysis by the Non-Heme Iron(II) Histone Demethylase PHF8 Involves Iron Center Rearrangement and Conformational Modulation of Substrate Orientation.
Chaturvedi SS, Ramanan R, Lehnert N, Schofield CJ, Karabencheva-Christova TG, Christov CZ. Chaturvedi SS, et al. ACS Catal. 2020 Jan 17;10(2):1195-1209. doi: 10.1021/acscatal.9b04907. Epub 2019 Dec 11. ACS Catal. 2020. PMID: 31976154 Free PMC article.
References
- Semenza G. L. (2000) Genes Dev. 14 1983-1991. - PubMed
- Epstein A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., et al. (2001) Cell 107 43-54. - PubMed
- Bruick R. K. & McKnight, S. L. (2001) Science 294 1337-1340. - PubMed
- Jaakkola P., Mole, D. R., Tian, Y.-M., Wilson, M. I., Gielbert, J., Gaskell, S. J., von Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., et al. (2001) Science 292 468-472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases