The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior - PubMed (original) (raw)
The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior
Brigitte Dauwalder et al. Genes Dev. 2002.
Abstract
The Drosophila somatic sex-determination regulatory pathway has been well studied, but little is known about the target genes that it ultimately controls. In a differential screen for sex-specific transcripts expressed in fly heads, we identified a highly male-enriched transcript encoding Takeout, a protein related to a superfamily of factors that bind small lipophilic molecules. We show that sex-specific takeout transcripts derive from fat body tissue closely associated with the adult brain and are dependent on the sex determination genes doublesex (dsx) and fruitless (fru). The male-specific Doublesex and Fruitless proteins together activate Takeout expression, whereas the female-specific Doublesex protein represses takeout independently of Fru. When cells that normally express takeout are feminized by expression of the Transformer-F protein, male courtship behavior is dramatically reduced, suggesting that male identity in these cells is necessary for behavior. A loss-of-function mutation in the takeout gene reduces male courtship and synergizes with fruitless mutations, suggesting that takeout plays a redundant role with other fru-dependent factors involved in male mating behavior. Comparison of Takeout sequences to the Drosophila genome reveals a family of 20 related secreted factors. Expression analysis of a subset of these genes suggests that the takeout gene family encodes multiple factors with sex-specific functions.
Figures
Figure 1
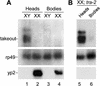
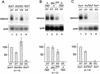
The takeout gene is expressed specifically in male heads and is derepressed in tra-2 mutant chromosomal females. (Top) Takeout expression in head and body RNA from nonstarved Canton-S male (XY) and female (XX) flies (A) and in tra-2PM6/ra-2PM7 mutant females (B) was examined by Northern analysis. The major band corresponding to takeout mRNA is indicated. The higher molecular weight band corresponds in size to takeout pre-mRNA. (Middle) Ribosomal protein 49 (rp49) hybridization to the same blot as a control for amount of RNA loaded. (Bottom) Hybridization with a probe for transcripts from the yolk protein 2 gene, which is expressed in female fat body.
Figure 2
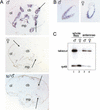
takeout RNA is expressed male specifically in brain-associated fat body, whereas expression in the antennae is not sex specific. (A,B) Frontal sections through male and female Canton-S heads were hybridized in situ with a takeout riboprobe originating from the 3′ untranslated region of the gene. Expression in the fat body (arrows) was observed in males, but not in females. Hybridization to the takeout probe was absent in takeout (to1) males. (B) Expression in the antennae was observed in both sexes. For reference, the positions of the central brain (cb), optical layers of the brain (ol), and mouthparts (mp) are indicated. (C) takeout RT–PCR on RNA from isolated antennae as well as whole male and female Canton-S flies is shown. Low-cycle PCR was performed with takeout and rp49 primers. The products were detected by Southern blotting and hybridization with an internal oligonucleotide probe.
Figure 3
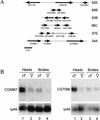
The Takeout family of proteins. An alignment is shown of the conserved protein-coding regions from the 20 different members of the takeout gene family encoded by Drosophila. Black shading indicates residues that are identical in at least eight members of the family, and gray shading indicates areas of similarity. CG7096-N and CG7096-C denote the N- and C-terminal sequences of the single ORF encoded by CG7096, in which the entire Takeout homology region is duplicated. In most cases, protein sequences are derived from translation of EST clones. In all other cases, sequences are based on the predicted genomic protein-coding regions as annotated by the Drosophila genome project (Adams et al. 2000). The sequences shown for CG14661, CG14457, and CG17279 are modified after splice junction reassignment on the basis of sequence alignment with other family members.
Figure 4
Multiple members of the takeout family are expressed sex specifically. (A) Organization of takeout family gene clusters are diagrammed. Arrows denote the length of each coding region and transcriptional orientation. Exons and introns are not indicated. Names of genes are below the arrows. The gray arrow denotes a gene (CG17189) that interrupts a cluster but is not in the takeout family. The cytological position of each cluster in the Drosophila genome is given at right. Three genes at dispersed locations (CG2645, CG14457, and CG13618) are not shown. (B) RNA blot hybridization analysis of CG5867 and CG7096 RNA expression in heads and bodies from males and females is shown. CG5867 is male enriched in both tissues, whereas CG7096 is male enriched only in heads. Hybridization of the same blot with rp49 mRNA is shown at bottom.
Figure 5
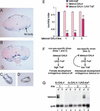
Feminization of Takeout-expressing cells disrupts male courtship behavior. GAL4 activity driven by the takeout promoter in sections from adult heads (A,B), antennae (C), and maxillary palps (D), was detected using a UAS–lacZ reporter gene. Frontal sections of takeout–GAL4/UAS–lacZ flies were stained with X-gal to detect β-galactosidase activity. Courtship indices of various males toward Canton-S virgin females are shown in E. Males carrying the UAS–traF (blue bar) or takeout–GAL4 (pink bars) transgenes individually have significantly higher courtship indices than the takeout–GAL4/UAS–traF males carrying both transgenes (red bars). The results from three different transgenic takeout–GAL4 lines are shown (lines 1,2,3). n = 10 for each group. ** indicates indices that were significantly different from those of parental strains (p < 0.001). (F) Diagram of the expected negative feedback loop set up in progeny from a cross of UAS–traF flies with the male-specific takeout–GAL4 line. (G) Northern analysis of endogenous takeout expression in takeout–Gal4/UAS–traF males and females as a measure for the feminization of _takeout_-expressing cells shows that takeout expression is drastically reduced in lines 1 and 2, but to a lesser degree in line 3. Expression of endogenous takeout RNA in the parental takeout–Gal4 adults from line 1 is shown for comparison (lanes 1,2).
Figure 6
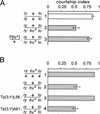
Mutations in takeout and fruitless interact to affect male courtship. Four sets of experiments are shown. (A) Courtship indices (± S.E.M.) of test males toward wild-type virgin females. The genotypes of males tested are indicated beside each bar. The different genotypes were generated by crossing various strains with ry506 to1 (lanes 1–4) or ry506 to+ (lanes 5–8). (Lane 1) takeout heterozygous males; (lane 2) homozygous mutant takeout males are not reduced in courtship; (lane 3) males homozygous for takeout and heterozygous for fru4 show a statistically significant reduction in courtship (marked by **, p < 0.001); (lane 4) males heterozygous for both takeout and fru4 show normal courtship. To control for a potential effect on courtship of the ry506 mutation on the takeout chromosome, parallel assays were performed on a ry506 strain that carries a wild-type takeout allele (to+ry; lanes 5–8). No effects of ry506 alone on courtship were observed. (B) Both the fru3 and fru4 allele interact with takeout to affect male courtship. ry phenotypes are indicated by white bars, ry+ phenotypes by dark bars. The fru4 and fru3 mutations are caused by independent P[ry+] insertion. n = 10 for each genotype. (C) The courtship index of to1 fru4 double homozygous mutant males toward females is lower than that of fru4 single mutant males alone (n = 19, p < 0.001). The courtship index of takeout single mutants is shown for comparison. (D) Males from a takeout1 strain that was outcrossed to the wild-type Canton-S strain [to1 (CS)] show a reduction in courtship (n = 9, p < 0.005).
Figure 7
Rescue of the to1/to1, _fru/_+ courtship defect by wild-type takeout or fruitless. (A) A genomic takeout+ transgene rescues the mutant phenotype (n = 8, p < 0.05). (B) Likewise, a duplication containing the wild-type fruitless gene (breakpoints at cytological locations 88D, 93D), rescues the courtship defect (lane 3; n = 6, p < 0.001), whereas a control duplication of similar origin (breakpoints at cytological locations 75D, 80) does not (lane 4; n = 6, p < 0.001).
Figure 8
Expression of takeout is affected by both doublesex and fruitless. RNA from whole flies was analyzed. (A) Northern analysis of takeout expression in dsx1 mutant flies shows that takeout expression in dsx1 mutant males is reduced (lane 2), and is derepressed in dsx1 females (lane 3) when compared with the expression in dsx1/+ siblings (lanes 1,4). (B) Forced expression of male-specific forms of dsx induce takeout in XX individuals. Females expressing the male form of dsx from the dominant mutation dsxSWE show activation of takeout (lane 3) compared with control females (lane 4). These males are dsxSWE/dsx+ and produce both Dsx-F and Dsx-M. There is no effect of dsxSWE/dsx+ on takeout expression in males (cf. lanes 1 and 2). (C) takeout expression is reduced in fru4/fru3 males (lane 2), but unaltered in fru4/fru3 females (lane 3) compared with their heterozygous siblings (lanes 1,4). (Bottom) Quantitation on the basis of several independent experiments (number indicated by n-value below each group). RNA levels were normalized to XY control males and rp49 controls.takeout expression in these males was assigned a value of 100 for each blot.
Similar articles
- A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain.
Drapeau MD, Radovic A, Wittkopp PJ, Long AD. Drapeau MD, et al. J Neurobiol. 2003 Apr;55(1):53-72. doi: 10.1002/neu.10196. J Neurobiol. 2003. PMID: 12605459 - The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song.
Rideout EJ, Billeter JC, Goodwin SF. Rideout EJ, et al. Curr Biol. 2007 Sep 4;17(17):1473-8. doi: 10.1016/j.cub.2007.07.047. Epub 2007 Aug 23. Curr Biol. 2007. PMID: 17716899 Free PMC article. - Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression.
Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, Yamamoto D. Usui-Aoki K, et al. Nat Cell Biol. 2000 Aug;2(8):500-6. doi: 10.1038/35019537. Nat Cell Biol. 2000. PMID: 10934470 - The roles of fruitless and doublesex in the control of male courtship.
Dauwalder B. Dauwalder B. Int Rev Neurobiol. 2011;99:87-105. doi: 10.1016/B978-0-12-387003-2.00004-5. Int Rev Neurobiol. 2011. PMID: 21906537 Review. - From fruitless to sex: On the generation and diversification of an innate behavior.
Peng Q, Chen J, Pan Y. Peng Q, et al. Genes Brain Behav. 2021 Nov;20(8):e12772. doi: 10.1111/gbb.12772. Epub 2021 Oct 21. Genes Brain Behav. 2021. PMID: 34672079 Review.
Cited by
- The mode of action of Fruitless: Is it an easy matter to switch the sex?
Sato K, Yamamoto D. Sato K, et al. Genes Brain Behav. 2020 Feb;19(2):e12606. doi: 10.1111/gbb.12606. Epub 2019 Sep 5. Genes Brain Behav. 2020. PMID: 31420927 Free PMC article. Review. - The Drosophila dopamine 2-like receptor D2R (Dop2R) is required in the blood brain barrier for male courtship.
Love CR, Gautam S, Lama C, Le NH, Dauwalder B. Love CR, et al. Genes Brain Behav. 2023 Feb;22(1):e12836. doi: 10.1111/gbb.12836. Epub 2023 Jan 13. Genes Brain Behav. 2023. PMID: 36636829 Free PMC article. - The circadian output gene takeout is regulated by Pdp1epsilon.
Benito J, Hoxha V, Lama C, Lazareva AA, Ferveur JF, Hardin PE, Dauwalder B. Benito J, et al. Proc Natl Acad Sci U S A. 2010 Feb 9;107(6):2544-9. doi: 10.1073/pnas.0906422107. Epub 2010 Jan 21. Proc Natl Acad Sci U S A. 2010. PMID: 20133786 Free PMC article. - Inducible Expression of Several Drosophila melanogaster Genes Encoding Juvenile Hormone Binding Proteins by a Plant Diterpene Secondary Metabolite, Methyl Lucidone.
Shin SW, Jeon JH, Kim JA, Park DS, Shin YJ, Oh HW. Shin SW, et al. Insects. 2022 Apr 29;13(5):420. doi: 10.3390/insects13050420. Insects. 2022. PMID: 35621756 Free PMC article. - doublesex functions early and late in gustatory sense organ development.
Mellert DJ, Robinett CC, Baker BS. Mellert DJ, et al. PLoS One. 2012;7(12):e51489. doi: 10.1371/journal.pone.0051489. Epub 2012 Dec 11. PLoS One. 2012. PMID: 23240029 Free PMC article.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The Genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
- An W, Wensink PC. Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes & Dev. 1995;9:256–266. - PubMed
- Baker B. Report of B. Baker. Drosophila Information Service. 1980;55:197.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases